
A Proposal for Explainable Breast Cancer Detection from Histological
Images
Lucia Lombardi
1
, Myriam Giusy Tibaldi
1
, Rachele Catalano
1
, Mario Cesarelli
2
, Antonella Santone
1
and Francesco Mercaldo
1
1
Department of Medicine and Health Sciences “Vincenzo Tiberio”, University of Molise, Campobasso, Italy
2
Department of Engineering, University of Sannio, Benevento, Italy
{francesco.mercaldo, antonella.santone}@unimol.it, mcesarelli@unisannio.it
Keywords:
Artificial Intelligence, Deep Learning, Digital Pathology, Breast Cancer.
Abstract:
Breast cancer is the most prevalent cancer among women globally, making early and accurate detection es-
sential for effective treatment and improved survival rates. This is the reason why, early and accurate breast
cancer detection is crucial for proper treatment planning to save a life. This paper presents a method designed
to detect and localize breast cancer using deep learning, specifically convolutional neural networks. The ap-
proach classifies histological images of breast tissue as either tumor-positive or tumor-negative. We utilize
several deep learning models, including a custom-built CNN, EfficientNet, ResNet50, VGG-16, VGG-19, and
MobileNet. Fine-tuning was also applied to VGG-16, VGG-19, and Mo bileNet to enhance performance.
The aim is to provide a more effective network, able to correctly detect and localise breast cancer, that could
support the physician in making clinical decisions. It could also prove to be a successful model to speed up
the diagnostic process and detect the possible presence of the disease at an early stage. Additionally, we in-
troduce a novel deep learning model called MR Net, aimed at providing a more accurate network for breast
cancer detection and localization, potentially assisting clinicians in making informed decisions. This model
could also accelerate the diagnostic process, enabling early detection of the disease. Furthermore, we propose
a method for explainable predictions by generating heatmaps that highlight the regions within tissue images
that the model focuses on when predicting a label, revealing the detection of benign, atypical, and malignant
tumors. We evaluate both the quantitative and qualitative performance of MR Net and the other models, also
presenting explainable results that allow visualization of the tissue areas identified by the model as relevant to
the presence of breast cancer.
1 INTRODUCTION
Breast cancer (BC) is the second most common can-
cer and the leading cause of cancer death among
women, after lung cancer. Currently, over 280,000
women are diagnosed with breast cancer each year in
the United States, and 44,000 die of the disease. De-
spite the enhancements in early detection and know-
ing of the molecular foundations of the biology of
BC, nearly 30% of the patients with “early-stage”
BC have disease recurrence. It is the uncontrolled
and irregular growth of breast tissues forming a lump
or tumor. These breast lesions are of two types:
benign and malignant. Diagnosis from a histolog-
ical image is the gold standard in diagnosing con-
siderable types of cancer. Histology allows to dis-
tinguish between normal tissue, non-malignant (be-
nign) and malignant lesions and to perform a prog-
nostic evaluation. Breast tissue biopsies allow pathol-
ogists to histologically assess the microscopic struc-
ture and elements of the tissue. Due to the com-
plexity and diversity of histology images, the man-
ual examination requires abundant knowledge and ex-
perience of the pathologists and is time-consuming
and error-prone. Therefore, Deep learning aims to
enhance accuracy and minimize human error, along-
side pathologists without replacing their role, foster-
ing a collaborative approach for improved diagnos-
tic outcomes. In this paper we propose, the descrip-
tion of convolutional neural networks (CNNs), capa-
ble of classifying the H&E stained breast histology
images into three classes: benign tissue, atypical le-
sions and malignant tumour.In particular, we consider
the following lesion types, Normal (N), Pathological
Benign (PB), Usual Ductal Hyperplasia (UDH), Flat
Epithelial Atypia (FEA), Atypical Ductal Hyperpla-
680
Lombardi, L., Tibaldi, M. G., Catalano, R., Cesarelli, M., Santone, A. and Mercaldo, F.
A Proposal for Explainable Breast Cancer Detection from Histological Images.
DOI: 10.5220/0013236500003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 680-685
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

sia (ADH), Ductal Carcinoma in Situ (DCIS) and In-
vasive Carcinoma (IC). The dataset on which to per-
form the experimental analysis was developed by the
collaboration of the National Cancer Institute IRCCS
‘Fondazione G. Pascale’ in Naples, the Institute for
High Performance Computing and Networks (ICAR)
and IBM Research in Zurich. The Dataset contains
4539 high-resolution histological images obtained by
applying hematoxylin and eosin (HE) staining and a
magnification factor of 40x. The images are all dif-
ferent sizes to ensure better heterogeneity of the sam-
ples. The results demonstrate the method ability to ac-
curately distinguish between three levels considered
(atypical tumour, benign and malignant.) and outper-
form other state-of-the-art methods based on feature
extraction. This approach has the potential to enhance
the computer-assisted diagnosis(CAD) of BC and im-
prove early diagnosis, contributing to the prevention
of avoidable deaths.
2 THE METHOD
This section shows the method we propose for BC de-
tection and localisation starting from tissue images.
We aim to find a model capable of classifying histo-
logical images as positive or negative for BC.
In detail, this is a multi-class classification prob-
lem because there are three classes to assign to a tis-
sue image under analysis, based on supervised learn-
ing. Clearly all the images in the training are already
labeled. The foundation of the methodology lies in
the selection of the dataset, selection of deep learn-
ing models, training and testing of these models, gen-
eration of explainability through Gradient-weighted
Class Activation Mapping (i.e., grad-CAMs) and
analysis of the results, as shown in figure 1.
Figure 1: The main steps of the proposed method.
2.1 Dataset and Preprocessing
The choice of dataset is fundamental, because it in-
fluences the performance, generalization, and relia-
bility of models. In the following case study, the
BReAst Carcinoma Subtyping (BRACS) (ICAR, Is-
tituto di Calcolo e Reti ad Alte Prestazioni, ) dataset
was adopted, consisting of histological images stained
with hematoxylin and eosin. This dataset was cho-
sen for the large number of images and the inclusion
of not only normal and cancerous images, but also
two atypical lesions, known as precancerous lesions.
(Sukhadia et al., 2023)
In particular, the types of lesions present in this
dataset are: Normal (N), Pathological Benign (PB),
Usual Ductal Hyperplasia (UDH), Flat Epithelial
Atypia (FEA), Atypical Ductal Hyperplasia (ADH),
Ductal Carcinoma in Situ ( DCIS) and invasive carci-
noma (IC). To optimize the dataset, a pre-processing
phase was carried out in order to obtain not only a
greater number of images, but also their more ho-
mogeneous distribution between the different classes.
The dataset utilized in this study presents three main
classes: atypical, malignant and benign. In particu-
lar, the atypical class includes images related to Flat
Epithelial Atypia (FEA) and Atypical Ductal Hyper-
plasia (ADH); the malignant class includes images of
Ductal Carcinoma in Situ (DCIS) and Invasive Car-
cinoma (IC) and the benign class includes images la-
beled as Normal (N), Pathological Benign (PB) and
Usual Ductal Hyperplasia (UDH). Subsequently a re-
sizing was carried out in order to obtain a size of
500x500 pixels for each image. To increase the num-
ber of examples to be provided to the deep learning
models, data augmentation was applied, in particular
the horizontal flip, brightness and zoom techniques.
(Gonz
´
alez-Castro et al., 2023)
Following this pre-processing phase, the final
dataset used contains 5628 images of which 80%
were allocated to training, 10% to testing and another
10% to validation, obtaining the following subdivi-
sion:
• training set: 4500 images of which 1500 classi-
fied as benign, 1500 as atypical and 1500 as ma-
lignant.
• validation set: 564 images of which 188 classified
as benign, 188 as atypical and 188 as malignant.
• test set: 564 images of which 188 classified as
benign, 188 as atypical and 188 as malignant.
2.2 The CNN Model
In this article we exploit the Standard CNN net-
work, created by the authors and the following CNNs
(He et al., 2024; Huang et al., 2024; Pan and Xin,
2024) already present in the literature: EfficientNet,
ResNet50, VGG-16, VGG-19 and MobileNet. The
Standard CNN is a network characterized by 13 lay-
ers. The convolutional block has three Conv2D layers
based on the application of 32, 64 and 128 3x3 size fil-
ters and ReLu activation respectively, alternating with
three MaxPooling2D layers. While the classification
A Proposal for Explainable Breast Cancer Detection from Histological Images
681

block has three Dense layers of 512, 256 respectively
with ReLu activation and three neurons with SoftMax
activation, alternating with 0,5 Dropout layers, used
to regularize the network. This network leverages the
categorical crossentropy loss function as it is a multi-
class classification.
2.3 Training
Once we developed the CNN models, the models
were trained on the considered dataset, selecting spe-
cific hyperparameters. These hyperparameters in-
clude the number of epochs, batch size and learning
rate. The values that led to obtaining better results
selected during the training phase are summarized in
the table 1.
2.4 Fine-Tuning
Lastly, we also implemented three additional models
using fine-tuning. Fine-tuning is a transfer learning
technique involving the use of a pre-trained model,
typically on a large dataset, we can adapt to our spe-
cific problem by continuing the training only for cer-
tain layers. Fine tuning requires two main steps: fea-
ture extraction, a phase involving the implementation
and training of a new classifier, and actual fine-tuning,
in which some of the layer that are closer to the clas-
sifier are unfrozen and re-trained. According to the
layer names adopted in Keras models, we unfroze:
• for MobileNet, the weights of the layers in
the last two convolutional blocks, starting with
“conv dw 12”;
• for VGG-16, the weights of all three layers
in the last convolutional block, starting with
“block5 conv1”.
• for VGG-19, the weights of all four layers
in the last convolutional block, starting with
“block5 conv1”.
2.5 Grad-CAM
Gradient-weighted Class Activation Mapping (Grad-
CAM) is a technique utilized in the field of deep learn-
ing to analyze the decisions made by CNNs in image
classification tasks (Zhou et al., 2023; Brunese et al.,
2022a; Brunese et al., 2022b; Martinelli et al., 2022;
Di Giammarco et al., 2023; Mercaldo et al., 2024;
Di Giammarco et al., 2024). Essentially, it reveals
which regions of an image capture the network’s at-
tention during predictions, thereby enhancing the un-
derstanding of the model’s decisions. Grad-CAM is
typically exploited for interpretability, as a matter of
fact deep neural networks are often treated as black
boxes due to their complex architectures. Grad-CAM
provides insight into their decision-making process
by highlighting which parts of an image are important
for a particular prediction. Moreover, it can be use-
ful for model debugging i.e., tt helps in understanding
and debugging model errors. By visualizing the re-
gions of an image that contribute most to a particular
prediction, researchers can identify potential biases or
misclassifications. Grad-CAM can also provide trust
and transparency, as a matter of fact in critical ap-
plications like healthcare or autonomous driving, it is
crucial to understand why a model makes a certain de-
cision. Grad-CAM enhances the trustworthiness and
transparency of AI systems by providing interpretable
explanations for their outputs.
3 EXPERIMENTAL ANALYSIS
In this section, we present the results of our experi-
mental analysis aimed at proposing a reliable method
for the detection and localization of BC. Specifically,
we analyze the metrics and confusion matrices ob-
tained during the classification phase to conduct a
quantitative analysis. Subsequently, we perform a
qualitative analysis by presenting images generated
via Grad-CAM to assess the features on which the
classification decisions were based. The results per-
tain to the classification performed using the images
from the test set described in section 2.1.
3.1 Quantitative Analysis
Table 2 shows the results of the experimental analysis
with the hyper-parameters given in Table 1.
Based on these metrics, it is determined that Mo-
bileNet and VGG-19 achieved the most favorable re-
sults among the evaluated networks, with an accu-
racy of 73%, a very satisfactory result considering
that other studies done on the same dataset achieved
an accuracy of 56 % (Brancati et al., 2022) and 66%
(Ahmed et al., 2023).
Table 3 shows the results of the experimental anal-
ysis carried out using fine-tuning. We chose to use
this technique only for the top three models, namely
MobileNet, VGG-16 and VGG-19, as can be seen
above.
Surprisingly, the accuracy achieved with this
method was slightly lower than the one achieved with-
out fine-tuning. The VGG-19 model exhibits the
worst results since it only reached an accuracy of 71%
against the 73% of the previous evaluation phase.
Along with the metrics, we also considered the
HEALTHINF 2025 - 18th International Conference on Health Informatics
682
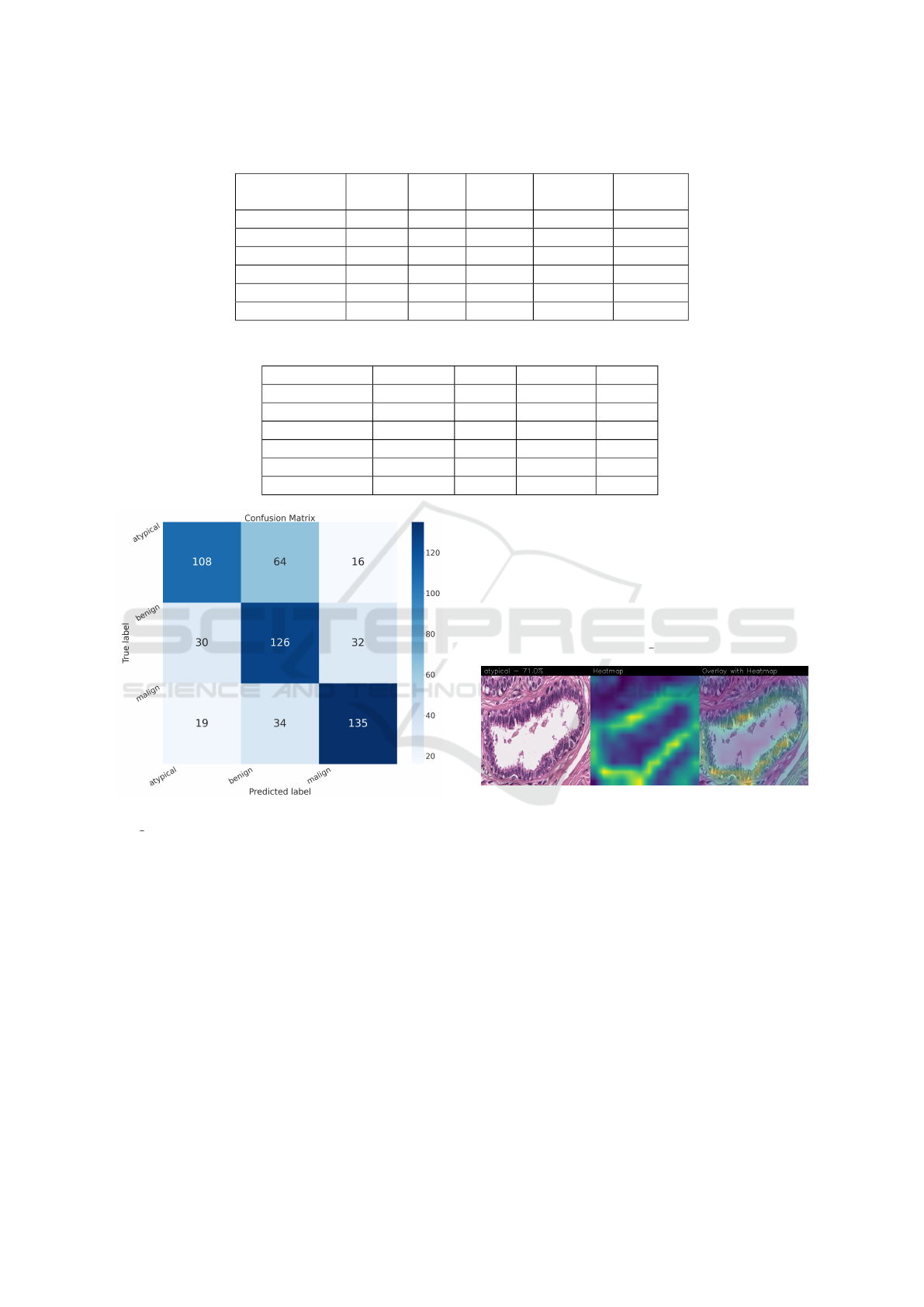
Table 1: Hyper-parameters selected during experimentation.
Model
Image
size
Batch Epochs
Learning
rate
Ex. time
Standard CNN 110×3 32 20 0.0001 0:08:57
EfficientNet 224×3 32 20 0.00001 1:21:48
ResNet50 110×3 32 50 0.0001 2:24:58
VGG-16 224×3 32 50 0.00001 15:44:49
VGG-19 224×3 32 50 0.00001 18:40:30
MobileNet 110×3 32 20 0.001 0:13:08
Table 2: Results of the experimental analysis.
Model Accuracy Loss Precision Recall
EfficientNet 0.6738 0.9003 0.6875 0.6631
ResNet50 0.7163 1.5485 0.7163 0.7163
VGG-16 0.7269 1.3160 0.7337 0.7181
VGG-19 0.7305 1.4156 0.7338 0.7234
MobileNet 0.7305 1.5697 0.7351 0.7234
Standard CNN 0.6737 0.9034 0.6824 0.6401
Figure 2: Confusion matrix obtained with the Stan-
dard CNN model.
confusion matrices in order to evaluate the classifi-
cation qualuty of the networks.
3.2 Qualitative Analysis
Drawing our conclusion merely on metrics would
lead us to consider the network as a black box, while
we want to propose a method that can be explain-
able to boost adoption of deep learning in real-world
medical activity. For this purpose, we also refer to
the images obtained through Grad-CAM, a technique
that proves to be extremely valuable for Explainable
Artificial Intelligence (XAI). The generated images
present in fact a heat-map that visually highlights the
areas the model relied on to make its decisions. (Ade-
biyi et al., 2024) This way we can understand more
thoroughly the reasons behind the classifica tion car-
ried out by a machine learning model. A heat-map
conveys informa tion through a color scale; specifi-
cally, in the images shown below, signifi cant regions
are represented in yellow, while less important areas
exhibit a blue/violet color. Below are the Grad-CAMs
obtained from the Standard CNN model:
Figure 3: Heatmap related to atypical cancer, correctly clas-
sified with a confidence of 71.0%.
In the case of atypical category (a precancerous
condition of the breast) the classifier utilizes the area
where the breast duct walls are darker purple in color.
Those walls are a little too thick with an excessive
number of cells, since epithelial atypica can grow to
a thickness of 5 or 6 cubic epithelial cells, as opposed
to the normal thickness of the breast duct lining of
about 2 cells. In fact, epithelial atypica is a pro lif-
eration of epithelial cells in the terminal duct-lobular
units (TDLU) of the breast. The cells are clustered in
acini that have rigid contours, round nuclei and even
chromatin and the cell borders are readily ap preci-
ated, creating the impression of a mosaic pattern. Se-
cretions and calcifications are present in the acinar lu-
mens.
A Proposal for Explainable Breast Cancer Detection from Histological Images
683
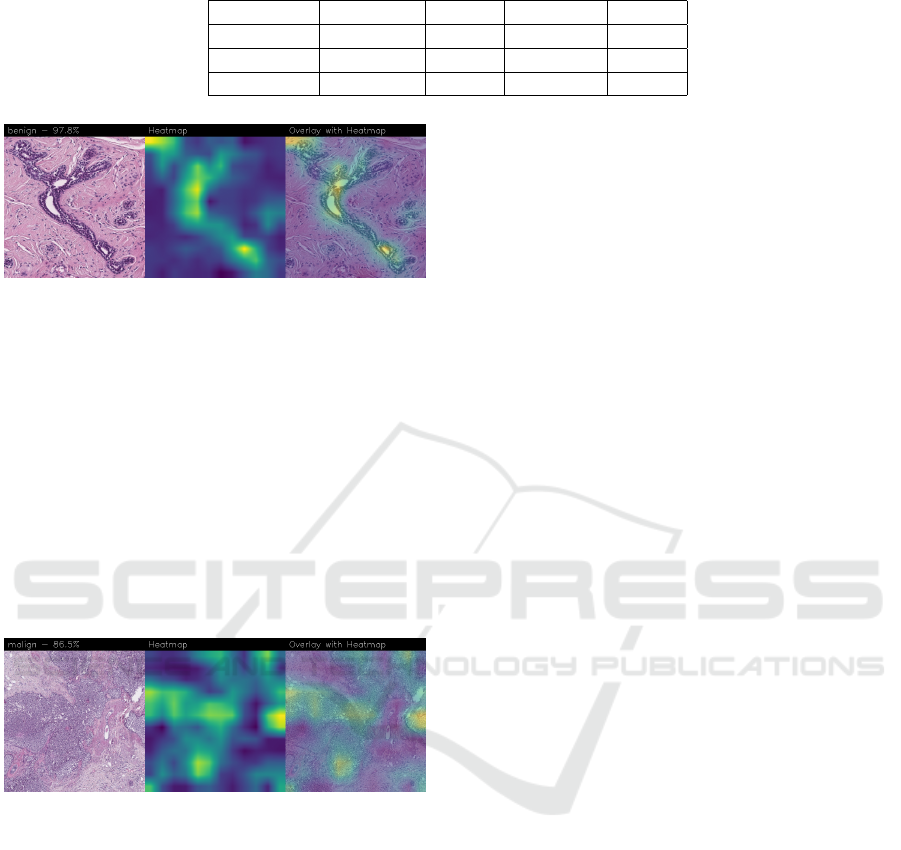
Table 3: Results of the experimental analysis using fine-tuning.
Model Accuracy Loss Precision Recall
MobileNet 0.7270 0.7132 0.7589 0.7198
VGG-16 0.7234 0.8434 0.7431 0.7181
VGG-19 0.7145 1.1417 0.7171 0.7057
Figure 4: Heatmap related to benign cancer, correctly clas-
sified with a confidence of 97.8%.
In the case of benign category the classifier de-
tects the area of normal tissue, consisting of glan-
dular tissue and adipose tissue. Ducts, lobules and
acini of the mammary gland are lined with epithelial
cells and immersed in adipose tissue. The model fo-
cuses on areas of the image containing the fibroade-
noma, a benign pathological nodule, that results from
the proliferation of the glandular epithelium and fi-
brous stroma of the breast. It is characterised by a fi-
broblastic stroma with glandular structures with cys-
tic spaces, surrounded by connective tissue forming
an enveloping capsule.
Figure 5: Heatmap related to malign cancer, correctly clas-
sified with a confidence of 86.5%.
In the case of malignant category the classifier re-
lies on large areas of the image, characterised by un-
differentiated malignant tissue, in which the tumour
cells have lost all their specific, normal histological
features and are therefore difficult to classify. In fact,
it is an invasive carcinoma.
4 CONCLUSION AND FUTURE
WORK
Accurate histopathological diagnosis is crucial for BC
as patient numbers surge and pathologist resources
dwindle. We believe that our study significantly im-
pacts the early diagnosis and identification of breast
cancer tumors and their subtypes, especially atypi-
cal and malignant tumors, thus improving patient out-
comes and reducing patient mortality rates. Although
the proposed model does not outperform state-of-the-
art models in terms of BC detection, it does in terms
of explainability, as the heat-maps generated using
Grad-CAM reveal a proper detection of the presence
of benign, atypical and malignant tumours. Both our
networks (Standard CNN e MR Net) base their de-
cision on the geometry of the structures, the number
and shape of the cells. However, the MR NET man-
ages to obtain more defined contours for the area of
interest, despite presenting a slight lower level of con-
fidence. Neither already existing models nor the fine-
tuned ones seem to reach these results for the Grad-
CAMs. It is clear that these models do not evaluate
the correct areas of the images, thus partially invali-
dating their results. Integrating AI into routine pathol-
ogy practice stands to improve diagnostic accuracy,
thereby contributing to reducing avoidable errors. De-
spite the existing hurdles, AI’s multifaceted contri-
butions to BC pathology hold great promise, provid-
ing enhanced accuracy, efficiency, and standardiza-
tion. Continued research and innovation are crucial
for overcoming obstacles and fully harnessing AI’s
transformative capabilities in breast cancer diagnosis
and assessment. From the future work point of view,
we will explore the possibility of considering other
models, for instance, related to object detection, to
understand whether it is possible to improve the per-
formance obtained in terms of BC localisation.
ACKNOWLEDGEMENTS
This work has been partially supported by EU DUCA,
EU CyberSecPro, SYNAPSE, PTR 22-24 P2.01 (Cy-
bersecurity) and SERICS (PE00000014) under the
MUR National Recovery and Resilience Plan funded
by the EU - NextGenerationEU projects, by MUR
- REASONING: foRmal mEthods for computA-
tional analySis for diagnOsis and progNosis in imag-
ING - PRIN, e-DAI (Digital ecosystem for inte-
grated analysis of heterogeneous health data related
to high-impact diseases: innovative model of care
and research), Health Operational Plan, FSC 2014-
2020, PRIN-MUR-Ministry of Health, the National
Plan for NRRP Complementary Investments D
∧
3
HEALTHINF 2025 - 18th International Conference on Health Informatics
684

4 Health: Digital Driven Diagnostics, prognostics
and therapeutics for sustainable Health care, Pro-
getto MolisCTe, Ministero delle Imprese e del Made
in Italy, Italy, CUP: D33B22000060001, FORE-
SEEN: FORmal mEthodS for attack dEtEction in au-
tonomous driviNg systems CUP N.P2022WYAEW,
ALOHA: a framework for monitoring the physical
and psychological health status of the Worker through
Object detection and federated machine learning, Call
for Collaborative Research BRiC -2024, INAIL, and
by Fondazione Intesa SanPaolo Onlus in the “Doctor-
ates in Humanities Disciplines” for the “Artificial In-
telligence for the Analysis of Archaeological Finds”
topic.
REFERENCES
Adebiyi, M. O., Olaniyan, D., Adebiyi, A. A., Olaniyan,
J., Amrevuawho, O. F., et al. (2024). Random
forest-based approach for integrating blood profile in
metastatic breast cancer classification. In 2024 In-
ternational Conference on Science, Engineering and
Business for Driving Sustainable Development Goals
(SEB4SDG), pages 1–6. IEEE.
Ahmed, F., Abdel-Salam, R., Hamnett, L., Adewunmi,
M., and Ayano, T. (2023). Improved breast cancer
diagnosis through transfer learning on hematoxylin
and eosin stained histology images. arXiv preprint
arXiv:2309.08745.
Brancati, N., Anniciello, A. M., Pati, P., Riccio, D., Scog-
namiglio, G., Jaume, G., De Pietro, G., Di Bonito, M.,
Foncubierta, A., Botti, G., et al. (2022). Bracs: A
dataset for breast carcinoma subtyping in h&e histol-
ogy images. Database, 2022:baac093.
Brunese, L., Brunese, M. C., Carbone, M., Ciccone, V.,
Mercaldo, F., and Santone, A. (2022a). Automatic
pi-rads assignment by means of formal methods. La
radiologia medica, pages 1–7.
Brunese, L., Mercaldo, F., Reginelli, A., and Santone, A.
(2022b). A neural network-based method for respira-
tory sound analysis and lung disease detection. Ap-
plied Sciences, 12(8):3877.
Di Giammarco, M., Dukic, B., Martinelli, F., Cesarelli, M.,
Ravelli, F., Santone, A., and Mercaldo, F. (2024). Re-
liable leukemia diagnosis and localization through ex-
plainable deep learning. In 2024 Fifth International
Conference on Intelligent Data Science Technologies
and Applications (IDSTA), pages 68–75. IEEE.
Di Giammarco, M., Mercaldo, F., Zhou, X., Huang, P., San-
tone, A., Cesarelli, M., and Martinelli, F. (2023). A
robust and explainable deep learning method for cer-
vical cancer screening. In International Conference
on Applied Intelligence and Informatics, pages 111–
125. Springer.
Gonz
´
alez-Castro, L., Ch
´
avez, M., Duflot, P., Bleret, V.,
Martin, A. G., Zobel, M., Nateqi, J., Lin, S., Pazos-
Arias, J. J., Del Fiol, G., et al. (2023). Machine
learning algorithms to predict breast cancer recurrence
using structured and unstructured sources from elec-
tronic health records. Cancers, 15(10):2741.
He, H., Yang, H., Mercaldo, F., Santone, A., and Huang, P.
(2024). Isolation forest-voting fusion-multioutput: A
stroke risk classification method based on the multidi-
mensional output of abnormal sample detection. Com-
puter Methods and Programs in Biomedicine, page
108255.
Huang, P., Li, C., He, P., Xiao, H., Ping, Y., Feng, P., Tian,
S., Chen, H., Mercaldo, F., Santone, A., et al. (2024).
Mamlformer: Priori-experience guiding transformer
network via manifold adversarial multi-modal learn-
ing for laryngeal histopathological grading. Informa-
tion Fusion, 108:102333.
ICAR, Istituto di Calcolo e Reti ad Alte Prestazioni. Bracs:
Breast carcinoma subtyping. https://www.bracs.icar.
cnr.it/.
Martinelli, F., Mercaldo, F., and Santone, A. (2022). Wa-
ter meter reading for smart grid monitoring. Sensors,
23(1):75.
Mercaldo, F., Di Giammarco, M., Ravelli, F., Martinelli, F.,
Santone, A., and Cesarelli, M. (2024). Alzheimer’s
disease evaluation through visual explainability by
means of convolutional neural networks. Interna-
tional Journal of Neural Systems, 34(2):2450007–
2450007.
Pan, H. and Xin, L. (2024). Fdts: A feature disentangled
transformer for interpretable squamous cell carcinoma
grading. IEEE/CAA Journal of Automatica Sinica,
12(JAS-2024-1027).
Sukhadia, S. S., Muller, K. E., Workman, A. A., and Na-
garaj, S. H. (2023). Machine learning-based predic-
tion of distant recurrence in invasive breast carcinoma
using clinicopathological data: a cross-institutional
study. Cancers, 15(15):3960.
Zhou, X., Tang, C., Huang, P., Tian, S., Mercaldo, F., and
Santone, A. (2023). Asi-dbnet: an adaptive sparse
interactive resnet-vision transformer dual-branch net-
work for the grading of brain cancer histopathological
images. Interdisciplinary Sciences: Computational
Life Sciences, 15(1):15–31.
A Proposal for Explainable Breast Cancer Detection from Histological Images
685
