
CFC Annotator: A Cluster-Focused Combination Algorithm
for Annotating Electronic Health Records by
Referencing Interface Terminology
Shuxin Zhou
1a
, Hao Liu
2
b
, Pritam Sen
1c
, Yehoshua Perl
1d
and Mahshad Koohi H. Dehkordi
1e
1
Department of Computer Science, New Jersey Institute of Technology, University Heights, Newark, U.S.A.
2
Department of Computer Science, Montclair State University, 1 Normal Ave, Montclair, U.S.A.
Keywords: Text Annotation, EHR, Cluster-Focused Combination, Dynamic Programming, Interface Terminology.
Abstract: In this paper, we present a novel algorithm designed to address the challenge of annotating electronic health
record (EHR) text using an interface terminology dataset. Annotated text datasets are essential for the
continued development of Large Language Models (LLMs). However, creating these datasets is labor-
intensive and time-consuming, highlighting the urgent need for automated annotation methods. Our proposed
method, the Cluster-Focused Combination (CFC) Algorithm, which stores intermediate results to minimize
annotation loss from terminology-based annotators, such as BioPortal’s (mgrep), while achieving high
coverage and significantly improving execution efficiency. We conduct a thorough evaluation of CFC on the
benchmark dataset MIMIC-III, using the previously developed Cardiology Interface Terminology (CIT).
Results show that CFC captured approximately 5,756 missed annotations from the baseline BioPortal (mgrep)
while achieving a remarkable improvement in execution speed across different size of datasets. These findings
demonstrate CFC’s scalability and robustness in processing large datasets, offering an efficient solution for
EHR text annotation. This work contributes to the preparation of large, high-quality training datasets for
Natural Language Processing (NLP) tasks in biomedical domains.
1 INTRODUCTION
The field of healthcare has undergone significant
transformation in recent decades, driven by the
widespread adoption and application of Electronic
Health Records (EHRs) (Blumenthal, 2009). EHRs,
which include diagnoses, radiology descriptions,
discharge summaries, and lab reports, are particularly
valuable as they contain up-to-date, patient-specific
information. However, despite their importance in
detailing individual patient conditions, much of this
information is recorded as unstructured text, often
using highly specialized clinical jargons.
To enable interoperability and enhance
healthcare quality through post hoc research, it is
critical to annotate these records with concepts from
standardized terminologies. Without such
a
https://orcid.org/0000-0003-1551-2781
b
https://orcid.org/0000-0002-1975-1272
c
https://orcid.org/0000-0003-2482-0057
d
https://orcid.org/0000-0003-1940-9386
e
https://orcid.org/0000-0003-3489-0892
annotations, the text remains ambiguous, vague, and
unsuitable for automated processing. Annotated data
not only enhances information sharing but also
empowers downstream applications, including
clinical decision support systems, disease
surveillance, and the development of cutting-edge
Large Language Models (LLMs).
Numerous annotation tools have been developed
to extract information from biomedical literature and
EHRs. Some of the most widely used tools in the
biomedical domain include MetaMap (Aronson &
Lang, 2010), cTAKES, and the NCBO Annotator:
1) MetaMap (Demner-Fushman et al., 2017):
Developed by the National Library of
Medicine (NLM), MetaMap maps concepts
extracted from biomedical and clinical text
to the Unified Medical Language System
Zhou, S., Liu, H., Sen, P., Perl, Y. and Dehkordi, M. K. H.
CFC Annotator : A Cluster-Focused Combination Algorithm for Annotating Electronic Health Records by Referencing Interface Terminology.
DOI: 10.5220/0013244500003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 195-206
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
195

(UMLS) (Bodenreider, 2004).
2) cTAKES: The Clinical Text Analysis and
Knowledge Extraction System combines
rule-based and machine learning techniques
to extract and normalize clinical concepts,
also for integration into UMLS (Savova &
others, 2010).
3) NCBO Annotator: This tool provides
broader functionality by mapping concepts
from text to any ontology hosted on
BioPortal (Jonquet et al., 2009)(Musen et
al., 2008). Its back-end Linux application,
Mgrep, efficiently matches text with billions
of ontology terms.
Ontologies, such as SNOMED CT (Donnelly,
2006), play a vital role in capturing meaningful
knowledge and representing relationships between
medical concepts (M & Chacko, 2020). Ontologies
and reference terminologies are designed to represent
conceptual knowledge in medicine and are not
optimized for application purposes (Rosenbloom &
others, 2006). For practical applications, interface
terminologies -- tailored specifically for specific
application -- are typically employed (Rosenbloom &
others, 2006)(Kanter et al., 2008)(Rosenbloom &
others, 2008).
In our prior research (V. Keloth et al., 2020)(V. K.
Keloth et al., 2023)(Dehkordi & others, 2023), we
developed two interface terminologies for COVID-19
and cardiology, leveraging the NCBO Annotator
during the iterative process of mining fine-grained
concepts. While NCBO Annotator performed well in
identifying seed phrases, they observed a key
limitation: it struggled to accurately annotate the
longest relevant text chunks. This occurred because
the tool indiscriminately identified all potential
concepts in the EHR text, leaving overlapping
matches unresolved.
To address this issue, we propose a Cluster-
Focused Combination (CFC) Annotation algorithm.
This algorithm is designed to annotate EHR text using
interface terminologies while maximizing annotation
coverage. Optimized CFC incorporates a strategy
storing intermediate results to address limitations in
conventional annotation techniques, specifically
those relying on the BioPortal (mgrep) terminology-
based annotator (Noy & others, 2009). Traditional
methods often fall short in maximizing coverage and
can miss important concepts due to sequential
matching constraints. In contrast, our approach
optimizes coverage by dynamically clustering
relevant terms and efficiently matching them to text,
minimizing the risk of annotation loss while
maintaining reference to interface terminologies.
The annotation process is divided into two major
steps:
1) Phrase Matching: Mgrep (Dai, 2021) is
used to generate a list of all matched phrases
in the text based on a user-provided interface
terminology. For this study, we applied the
Cardiology Interface Terminology (CIT) (V.
Keloth et al., 2020)(Dehkordi & others,
2023).
2) Cluster-Focused Annotation: Using the
matched phrases list, the CFC annotation
algorithm identifies an optimal combination
of concepts for each sentence to ensure high-
coverage annotations in the EHR text.
Our approach is generic and can be applied to any
user-generated terminology, enabling robust and
comprehensive annotation of EHRs for various
applications with the optimal execution time.
2 BACKGROUNDS
2.1 MIMIC-III Dataset
MIMIC-III, short for Medical Information Mart for
Intensive Care version 3, is an open-sourced, de-
identified database that captures critical care data from
patients admitted to the intensive care units at Beth
Israel Deaconess Medical Canter in Boston,
Massachusetts (Saeed et al., 2002). This extensive
dataset includes a wide array of information such as
vital signs, medications, laboratory results, procedural
and diagnostic codes, as well as billing details.
2.2 Clinical Interface Terminology
Rosenbloom et al. (Rosenbloom & others,
2006)(Kanter et al., 2008)(Rosenbloom & others,
2008) describe that clinical interface terminologies
aid healthcare practitioners by enabling rapid
retrieval of patient information through a structured
set of healthcare-related terms. These terminologies
are specifically tailored for end-users, incorporating
commonly used clinical phrases and colloquial
expressions, in contrast to reference terminologies
that aggregate clinical data based on standardized
concepts.
In previous work (V. K. Keloth et al., 2023)(V.
Keloth et al., 2020)(Dehkordi & others, 2023), we
developed two pipelines for iteratively generating
fine-granular clinical phrases from the Mimic-III
cardiology notes and the Radiopeadia scan
descriptions by using concatenation and anchoring
HEALTHINF 2025 - 18th International Conference on Health Informatics
196
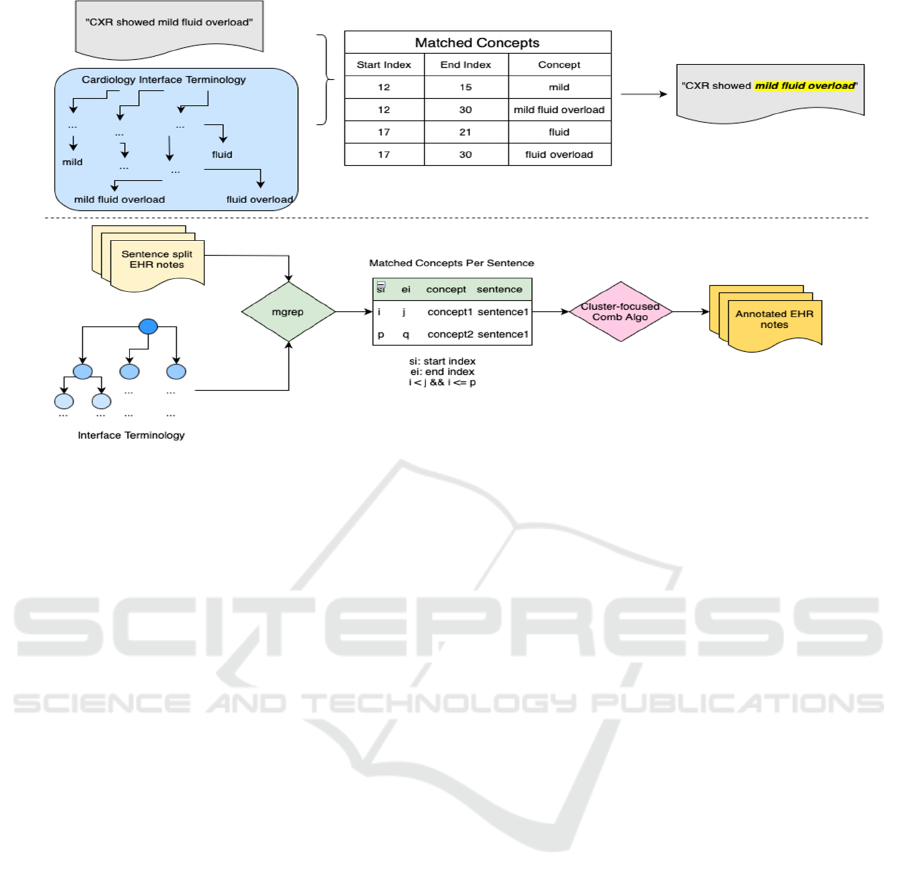
Figure 1: The entire workflow of the annotation pipeline.
operations. The generated concepts from these
processes were used to construct two distinct
interface terminologies: one for cardiology(V. Keloth
et al., 2020)(Dehkordi & others, 2023) and one for
Covid-19(V. K. Keloth et al., 2023).
2.3 NCBO BioPortal
The NCBO BioPortal is an open-source platform
maintaining a continually growing collection of 1,158
biomedical ontologies. These ontologies are available
in four different formats, including: 813 are in OWL
(Web Ontology Language) format (Dean et al., 2004),
106 in OBO (Open Biomedical Ontologies) format,
30 in the UMLS’s RRF format, and 122 in SKOS
(Simple Knowledge Organization System) format
(Miles & Bechhofer, 2009). BioPortal covers a broad
range of topics ranging from Biological Process, to
chemistry (Musen et al., 2008)(V. Keloth et al.,
2020).
The BioPortal also provides varied of services for
ontology users and allows them to upload their own
developed terminologies. Once successfully
submitting the terminology, the BioPortal generates
and displays additional statistics about the uploaded
terminology, including the number of classes and
properties.
2.4 Mgrep Text Matching Tool
Mgrep is a command line tool that enables users to
search through text files for lines that match a
specified regular expression (Dai, 2021). It is used as
the back-end algorithm for BioPortal Annotator
function. It functions similarly to the well-known
Unix tool, grep, but extends its capabilities by
supporting searches for patterns spanning multiple
lines. While mgrep is not designed to replace grep, it
is built to be compatible with it, meaning its options
and behaviours closely align with those of grep. In
our algorithm, we use mgrep to identify all relevant
concepts from CIT that match phrases in the text,
which are then used as candidates for our cluster-
focused combination algorithm to annotate the EHR
text.
3 METHOD
A user constructed interface terminology contains
plenty of phrases from the user provided text dataset:
some of them are high granular phrases, and some of
them are relatively short in the length. Annotating the
text with this set of phrases from the interface
terminology with our method can be illustrated in the
Figure 1.
Initially, we use mgrep to align text segments
with the Cardiology Interface Terminology (CIT) (V.
Keloth et al., 2020). Mgrep generates all matched
phrases including cases of overlapping concepts of
CIT.
Within the sentence, these matched phrases can
be classified into 5 different conditions of positioning
which will be discussed in section 3.1. Subsequently,
CFC Annotator: A Cluster-Focused Combination Algorithm for Annotating Electronic Health Records by Referencing Interface
Terminology
197

we apply a cluster-focused algorithm to determine the
optimal combination of concepts in each sentence and
highlight each concept in the text.
3.1 Five Conditions
Consider a sentence S of n characters S [0, n-1],
which is annotated by a terminology T. Suppose that
m occurrences of concepts of T were identified in the
sentence S. Let C
1
, C
2
, …, C
m
be the concepts
identified in S and stored in Dictionary D. Let L
i
and
Ri be the respective starting index and ending index
of C
i
in S, where L
i
≤ R
i
for i = 1, m. The concepts C
i
in S are ordered by their Li: (L
1
≤ L
2
… ≤ L
m
).
Example 1: Sentence S: “CXR showed mild fluid
overload.”
Table 1: Dictionary D associated to sentence S.
L(start) R(end) Concep
t
C
1
12 15 mild
C
2
12 30 mild fluid overloa
d
C
3
17 21 flui
d
C
4
17 30 fluid overloa
d
1. Two concepts C
j
and C
k
(j < k) in S are called
disjoint, if R
j
+ 1 < L
k
. (Plus 1 is necessary, since
it must have a space between C
j
and C
k
), for
examples:
a. Concept C
1
“mild” and C
4
“fluid
overload” are disjoint.
b. Concept C
1
“mild” and C
3
“fluid” are
disjoint
2. Two concepts C
j
and C
k
(j < k) in S are called left-
adjusted, if L
j
= L
k
and R
j
< R
k
. for example:
a. Concept C
3
“fluid” and C
4
“fluid
overload” are left-adjusted
3. Two concepts C
j
and C
k
(j < k) in S are called
right-adjusted, if L
j
< L
k
and R
j
= R
k
. See
examples:
a. Concept C
3
“mild fluid overload” and C
4
“fluid overload” are right adjusted
4. Two concepts C
j
and C
k
(j < k) in S are called
overlap, if L
j
< L
k
< R
j
< R
k
.
a. For example, supposing the two
concepts are "mild fluid overload" and
"fluid overload XXX" (XXX refers to a
word), then "fluid overload" is the
overlapping.
5. Two concepts C
j
and C
k
(j<k) in S satisfy that C
j
contains C
k
, if L
j
< L
k
and R
j
> R
k.
See examples:
a. Concept C
2
“mild fluid overload”
contains C
3
“fluid”.
By annotation of S using a terminology T, we
refer to a subset of Dictionary D, such that all C
i
in
the subset are mutually disjoint. When we consider an
option of annotation for sentence by the terminology
T, we have two purposes. The first purpose is to find
a set of concepts for annotating such that the total
number of words in those concepts is maximized.
This purpose will lead to a high value of the
annotation coverage for a given text. The other
purpose is capturing the semantics of the sentence as
intended by its author. Towards this goal, the CIT
includes high granularity concepts, which better
capture the semantic in the sentence. These concepts
are typically longer and contribute to higher breadth
of the annotation. The ultimate purpose is to serve
customers of the annotation which are trying to
comprehend the content of the text from its annotated
parts. For this purpose, our annotation is not only
matching the text to the existing terminology and
listing all the matched concepts which are possibly
redundant or overlapped, but we also output the text
with highlighted annotations in the format of html. It
is more user friendly.
Consider the case that all K concepts of T found
in S are mutually disjoint. In this case, all of them are
selected for the annotation, and the coverage of the
sentence is optimal. However, in most cases, we
encounter pairs of concepts which are left-adjusted,
right-adjusted, overlapping, or in containment
relation.
3.2 Basic Cluster-Focused
Combination (CFC) Algorithm
Considering the sentence: “The patient was taken on
to the operating room for a redo mitral valve surgery
with mitral valve replacement”. The phrases of the
sentences which are matched to the concepts in CIT
are listed in Table 2. We observe that the concepts
“patient” “operating room” and “redo” where isolated
concepts which did not interact with any other
concepts of the dictionary. Hence, those disjoint
concepts of the dictionary are selected for the
annotation because they will appear in any annotation
of the sentence using D. The challenge for selecting
concepts for the annotation appears when we have a
set of concepts with overlap. We see the clusters for
five concepts C
4
to C
8
and three concepts C
9
to C
11
in
Table 2. The challenge is how to pick a combination
of concepts for such clusters which optimizes in
coverage and breadth.
HEALTHINF 2025 - 18th International Conference on Health Informatics
198
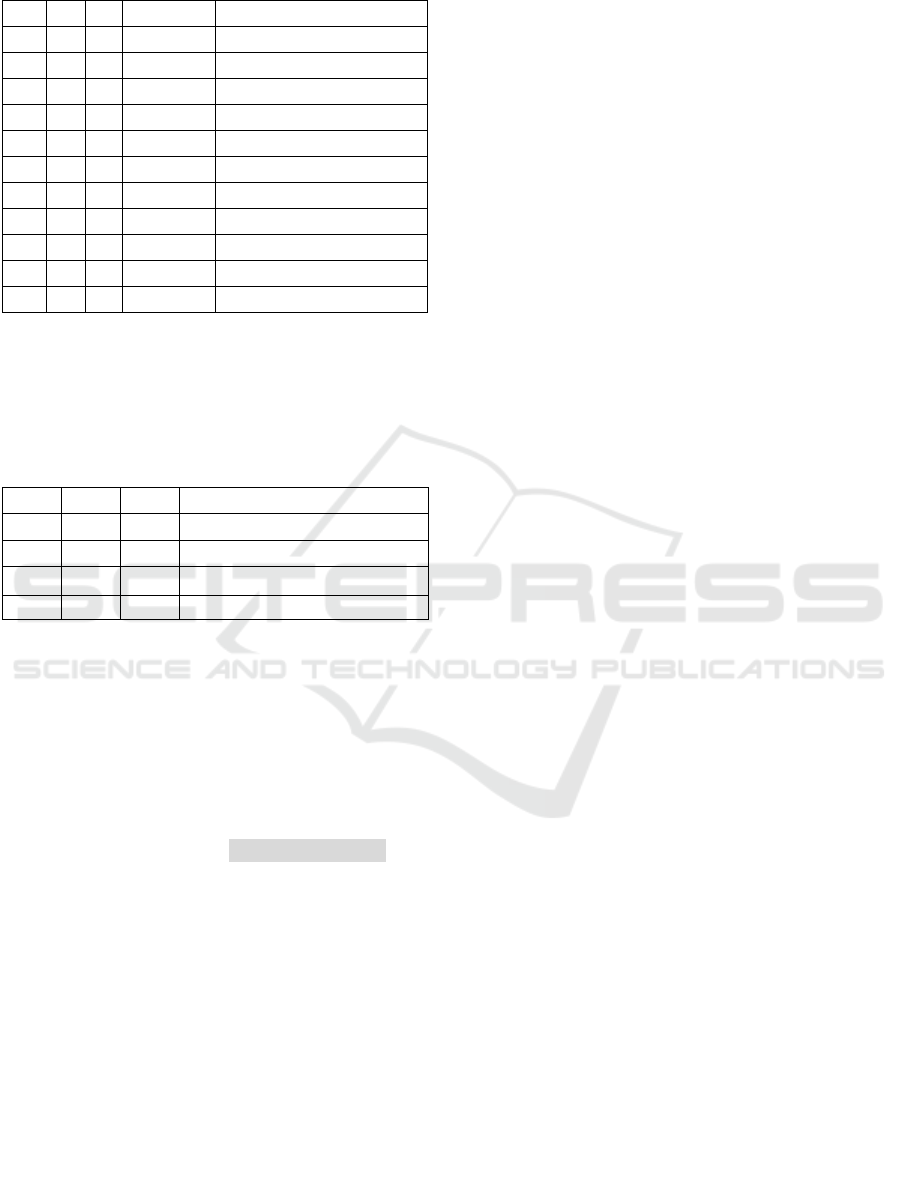
Table 2: Dictionary D associated to Example 1.
L R # of words Concept
C
1
5 11 1 patient
C
2
33 46 2 operating room
C
3
54 57 1 redo
C
4
59 70 2 mitral valve
C
5
59 78 3 mitral valve surgery
C
6
66 70 1 valve
C
7
66 78 2 valve surgery
C
8
71 78 1 surgery
C
9
85 96 2 mitral valve
C
10
92 96 1 valve
C
11
85 107 3 mitral valve replacement
Note*: L refers to start index, and R refers to end index. The reason
for not using S to represent start index is to distinguish with the
abbreviation of Sentence S.
In Example 2: “CXR showed mild fluid overload”, we
have the matched concepts as in Table 3.
Table 3: Matched concepts in Example 1.
L R Concept
C
1
12 15 mild
C
3
12 30 mild fluid overload
C
4
17 21 fluid
C
5
17 30 fluid overload
There are two combinations of concepts sharing
the maximum annotated words: COMB
1
= C
1
+ C
5
and COMB
2
= C
3
.
Since COMB
1
has two concepts and COMB
2
has
one concept, we select COMB
2
for annotation in S,
because there is COMB
2
contains only one concept.
When a tie between two combinations on the number
of words, the one contains less concepts is preferable
to increase the breadth. As a result, the sentence S is
annotated as: “CXR showed mild fluid overload”.
Before generating the COMB of the whole
sentence, it is necessary to recap the definition of
“cluster” in the algorithm. “Cluster” of a sentence. For
this, we define a graph where each annotated concept
is a node. Two nodes are connected by an edge if there
is an overlap between the phrases of the concepts
represented by the nodes. That is the concepts are left-
adjusted, right-adjusted or overlapping. A maximal
connected subgraph of the graph representing the
sentence is called a “cluster”. The purpose for finding
clusters of a sentence is: when a sentence contains a
cluster, a decision will be made on which concept(s)
should be selected in the cluster for annotating the
sentence. That is, which concept(s) should be
collected and kept in the COMB set.
3.3 Optimized Cluster-Focused
Combination (CFC) Algorithm
The algorithm for finding an annotation of a cluster
of k concepts with maximum coverage and breadth
scans the concepts of the cluster from left to right,
considering for each concept the solutions with and
without this concept. The complexity is O(N
k
). In
practices, we observed that for only 150 notes, it takes
more than half hour. Due to the issue of high time-
consuming, we optimize the algorithm by importing
a memorization mechanism.
3.3.1 Workflow of Annotation Process by
Using Optimized CFC
Like in the basic version of the Cluster-focused
Combination Algorithm, concepts are first sorted by
their start index. If two or more concepts share the
same start index, they are then sorted by their end
index in ascending order. A strategy is then applied to
determine whether each matched concept should be
selected or omitted from the solution. Based on the
current concept selection, there are five possible
scenarios:
1) If the Current Concept is selected, and when
the Next Concept overlaps with the Current
Concept, then, keep the Current Concept and
continue checking the subsequent concepts
to find one that doesn’t overlap.
2) If the Current Concept is selected, and when
the Next Concept does NOT overlap with
the Current Concept, then, select the Next
Concept and add to the Set.
3) If the Current Concept is selected, when the
Next Concept does NOT overlap with the
Current Concept, do not select the Next
Concept.
4) If the Current Concept is not selected, select
the Next Concept and add to the Set.
5) If the Current Concept is not selected, do not
select the Next Concept either.
In this project, we divide the problem into
smaller subproblems by considering subsets of
concepts starting from each concept. For each
concept, we maintain two scores: the maximum
number of words covered when (i) selecting the
concept and (ii) omitting it. Finally, after calculating
the scores for all concepts in reverse order, we
determine the result by checking the score for the
first concept, which represents the overall score for
the complete solution.
CFC Annotator: A Cluster-Focused Combination Algorithm for Annotating Electronic Health Records by Referencing Interface
Terminology
199

Figure 2: Example of the workflow by optimized CFC.
Figure 2 illustrates an example with six matched
concepts within a single sentence. In Figure 2, this
process is illustrated using a backwards dynamic
selection approach. For each concept, we represent a
pair as (a, b), where a and b denote the scores when
omitting and selecting the concept, respectively.
Starting with the last concept, C6, we assume only
this concept is available. If C6 is selected, the Current
Set becomes {{C6} Len:3}, with a length of 3 words;
if C6 is omitted, the set is empty, represented as {{}
Len:0} with a length of 0 words. Thus, for C6, we
store (0, 3), where 0 and 3 are the scores for omitting
and selecting C6, respectively.
Next, we consider C5, which overlaps with C6.
If we add C5 to the Current Set, we cannot select C6,
resulting in {{C5} Len:4}. By omitting C5, we retain
the option to select C6, which produces the set {{C6}
Len:3}. Since, for each concept, we store the
maximum score between selecting and omitting it, we
record (3, 4) for C5.
Moving to C4, which overlaps with both C5 and
C6, we find that selecting C4 requires omitting both
C5 and C6, resulting in a score of 5. By omitting C4,
we retain the highest score starting from C5, which is
4. Thus, we store (5, 4) as the scores for C4,
corresponding to the sets {{C4} Len:5} and {{C5}
Len:4}, respectively.
Continuing this process, if we select C3, we
cannot select C4, so we choose the best result from
C5, forming the set {{C3, C5} Len:6} (calculated as
{{C3, C5} Len:2+4}). C2 and C1 can be added
without any conflict, as they do not overlap with other
concepts, resulting in the set {{C1, C2, C3, C5}
Len:10}, calculated as {{C1, C2, C3, C5}
Len:3+1+6}.
Finally, the optimal score is achieved with C1,
which represents the best possible combination of
concepts. Thus, the best concept combination for the
annotation text is {C1, C2, C3, C5} with L=10.
3.3.2 Pseudo Code of Optimized CFC
In Algorithm 1, we present the pseudo-code for the
optimized (CFC) algorithm. This optimization
merges two tasks: "finding the maximum number of
words annotated" and "determining the best
combination of concepts"-- into a single function:
find_decide_best_comb_dp. This function is
organized into five key sections that guide the
decision-making process for selecting the best
concept combination at each stage.
The first two sections address the initialization of
the case base and previously traversed cases, laying
the groundwork for recursive exploration. Section 3
handles the scenario where the current concept is
“not” included in the solution. In this case, the
algorithm simply copies the previous state of results
to a list, not_take_curr_concept, for storage and
continues the recursion without modifying the
solution. If we decide to include the current concept
in the combination, Section 4 initiates two primary
operations. First, it locates a a next qualified concept
that does not overlap with the current concept; this
non-overlapping condition ensures that we maintain a
conflict-free solution as we proceed. Next, the current
concept’s index is added to
next_decision_concept_list, which accumulates the
indices of concepts chosen in the optimal
combination so far. At this point, we update the
take_curr_concept list by:
1. Updating the “Maximum Number of Words
Annotated” by adding the word count of the
current concept.
HEALTHINF 2025 - 18th International Conference on Health Informatics
200

Algorithm 1: Find the optimal combination of concepts for annotating the sentence.
2. Increasing the “Minimum Number of
Concepts” by one, as we add the current
concept to the optimal set.
3. Updating the best current concepts list to
reflect the newly added concept.
With not_take_curr_concept and
take_curr_concept lists now fully populated, we have
enough information to decide whether to include the
current concept.
Finally, Section 5 performs a comparison between
not_take_curr_concept and take_curr_concept lists
based on two prioritized criteria. First, it evaluates the
“number of words annotated” -- the decision that
annotates the maximum words is preferred. If there is
a tie in this criterion, it then considers the “number of
concepts”, preferring the option that includes fewer
concepts for efficiency. The "winning" combination at
this stage is stored in curr_comb, representing the best
possible combination up to the current point in
recursion. at this stage is stored in curr_comb,
representing the best possible combination up to the
current point in recursion.
CFC Annotator: A Cluster-Focused Combination Algorithm for Annotating Electronic Health Records by Referencing Interface
Terminology
201
4 RESULTS
We tested our algorithm on a corpus of de-identified
Cardiology Information notes from the Mimic-III
dataset. The objective is to annotate relevant terms
within the notes by referencing the CIT (V. Keloth et
al., 2020)(Dehkordi & others, 2023). For the
evaluation, we employed two metrics: coverage and
breadth. Coverage measures the percentage of
concepts from the CIT captured within the EHR
notes, reflecting the extent to which the interface
terminology comprehensively annotates the text.
Breadth, on the other hand, indicates the average
number of words per annotated concept, representing
the specificity or granularity of concepts within the
CIT. High coverage implies extensive text annotation
by the CIT, signifying its thoroughness, while breadth
highlights the conceptual detail included in the
terminology. The equations are as follows:
We selected the BioPortal Annotator and a
baseline Cluster-focused-Combination annotation
algorithm for comparison. Given that our annotation
approach relies on precise matching with the concepts
within a specialized domain-specific interface
terminology using large language models is not
suitable for this study. This focus on domain-specific,
deterministic concept matching ensures that the
annotations remain consistent with the defined
terminological scope of CIT, which is essential for
accurate representation and evaluation in this context.
4.1 Coverage and Breadth
The experiments were conducted on two datasets: a
small-scale text dataset annotated with an earlier
version of the Cardiology Interface Terminology
(CIT) and a large-scale dataset annotated with the
final version. For the smaller dataset, we randomly
selected 150 cases from MIMIC-III Event Notes and
annotated them using CIT_V
3.2
. For the larger dataset,
comprising 500 notes, we applied the final version,
CIT_V
5.2
, to evaluate the capacity of the annotators in
handling a larger volume of data.
These phrases are constructed by concatenating
and anchoring CIT_V
3.2
concepts. Consequently,
annotating the 500-note dataset with CIT_V
5.2
introduces a higher number of potential matched
phrases and more complex overlapping cases when
identifying concepts for annotating in the text. This
second dataset, therefore, presents a greater challenge
for the annotation process, testing the robustness and
accuracy of the annotators under more intricate
conditions.
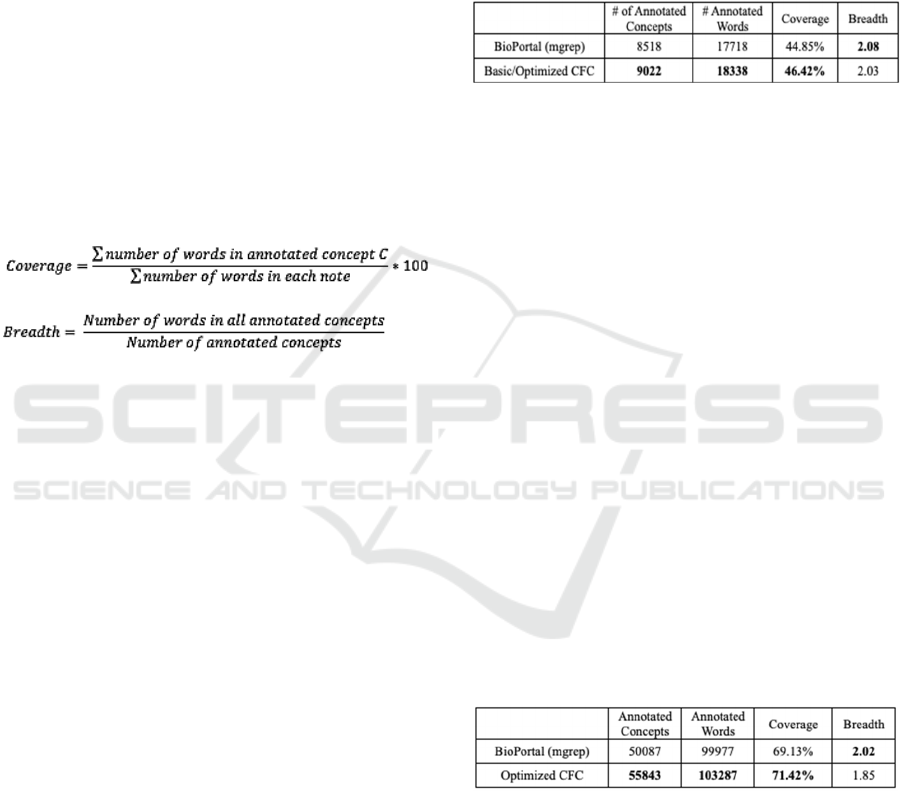
Table 4: The Comparison of # of Annotated Concepts,
Coverage and Breadth among Two Annotators on 150
Notes.
Table 4 shows that 504 concepts were missed by
the BioPortal (mgrep) annotator but were
successfully identified by the Cluster-Focused
Combination (CFC) algorithm. Although the dataset
is relatively small, the increase in coverage between
BioPortal (mgrep) and the CFC algorithm is modest.
The slightly lower breadth in CFC annotations
reflects two key points:
1) The generated interface terminology includes
highly granular concepts.
2) The BioPortal annotator prioritizes the longest
matching concept per sentence, often omitting
shorter, overlapping concepts. This approach
sacrifices some information by reducing the
text coverage.
This second limitation motivated the
development of the CFC algorithm, designed to
address these gaps by capturing both long and short
overlapping concepts. Additionally, the optimized
CFC algorithm shares core functionalities with the
basic CFC version, resulting in identical outcomes for
the number of annotations, coverage, and breadth.
Table 5: The Comparison of # of Annotated Concepts,
Coverage and Breadth among Two Annotators on 500
Notes.
Table 5 compares the performance of two
annotation methods on the larger 500-note dataset.
Here, the CFC algorithm captures 5,756 concepts
missed by BioPortal, a significant increase compared
to the 504 missed concepts in the 150-note dataset.
While the dataset size grew by 3.3 times, the number
of missed annotations increased approximately 11
time, highlighting the annotation or information loss
issue faced by BioPortal.
For the larger dataset, we did not include
HEALTHINF 2025 - 18th International Conference on Health Informatics
202
experiments with the basic CFC algorithm due to its
high time complexity, O (2
k
) per sentence,
where N=2 represents the number of matched
candidate concepts, and k is the size of the concept
combination. Detailed runtime analysis can be found
in section 4.2.
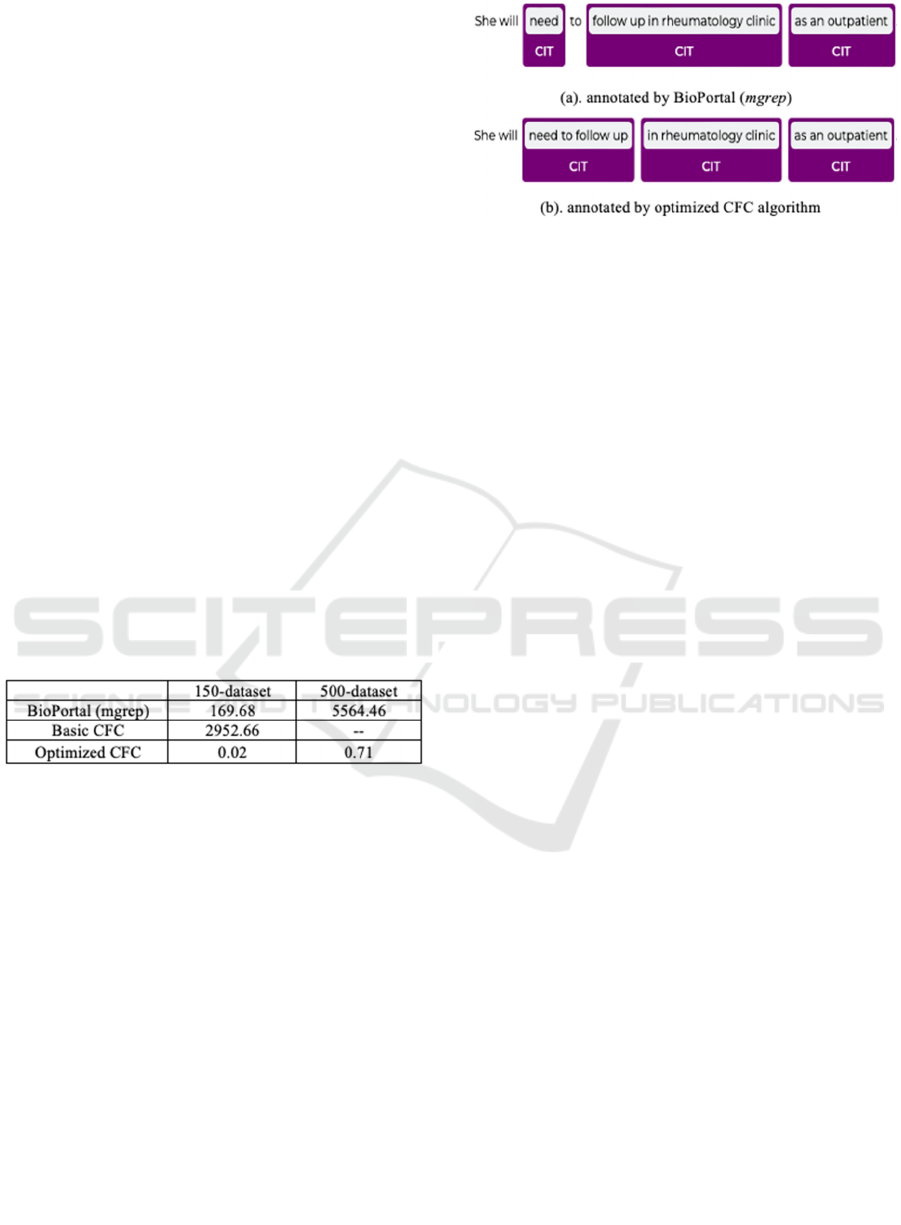
4.2 Runtime Analysis
The optimized CFC algorithm significantly enhances
the time efficiency of the basic CFC algorithm by
employing memorization techniques. Instead of
processing every possible combination of concepts,
the optimized CFC algorithm evaluates scores for all
subsequence in the sorted list of concepts. Therefore,
the theoretical time complexity is improved from
O(2
k
) to O(k
2
). As shown in Table 3, the runtime of
the basic CFC on the 150-note dataset is 2,952
seconds and for BioPortal with 500-notes which took
5564 seconds, meaning that is not efficient for
processing larger datasets. In contrast, the optimized
CFC drastically reduces execution time from hours to
milliseconds. Annotating the 500-note dataset with
the optimized CFC takes less than one second,
demonstrating its scalability and feasibility for
processing large biomedical datasets.
Table 6: The Comparison of Execution Runtime (seconds)
among Three Annotators on Two Datasets.
4.3 Case Study
Figure 6 presents an example for sentence “She will
need to follow up in rheumatology clinic as an
outpatient.”, annotated using both the BioPortal
(mgrep) and CFC algorithms. In Figure 3(a), the
BioPortal (mgrep) identifies and annotates the longest
matched phrase, “follow up in rheumatology clinic”.
The remaining two segments of the sentence are
annotated with non-overlapping concepts,
specifically “need” and “as an outpatient”.
Altogether, this approach annotates 9 words, covering
75% of the sentence. By contrast, the CFC algorithm
identifies three additional concepts within the
sentence: “need to follow up”, “in rheumatology
clinic”, and “as an outpatient”. Although CFC does
not select the longest matched concept, “follow up in
rheumatology clinic”, it annotates a total of 10 words
-- one more than BioPortal (mgrep).
Figure 3: Annotation for example one by BioPortal (mgrep)
and optimized CFC annotator.
In another instance, important information is
missed by the BioPortal (mgrep) annotator. Figure 4
illustrates a comparison between BioPortal (mgrep)
and CFC annotations for Example Two, the
sentence: “Although the POBA to the RCA lesion was
unsuccessful, the L Cx lesion was successfully stented
with a BMS.” In Figure 4, the BioPortal (mgrep)
misses the important information “BMS” in its
annotation, as it selects the concept “successfully
stented” over the shorter “successfully”. This choice
results in the omission of the important detail “BMS”.
In contrast, the CFC algorithm captures all relevant
information, providing a more comprehensive and
accurate annotation.
A similar issue arises in Figure 5 with the
sentence: “Had asymptomatic run of NSVT with
stable vital signs.” In Figure 5(a), the BioPortal fails
to identify the term “NSVT”. Instead, it
selects “asymptomatic run” as the annotated concept
due to its longer phrase length compared
to “asymptomatic”. However, “NSVT” appears
within another concept, “run of NSVT”, which
overlaps with “asymptomatic run”. This overlap
leads BioPortal to skip annotating “run of NSVT”,
resulting in the omission of “NSVT” from the
annotation.
5 DISCUSSIONS
Our Cluster-Focused Combination (CFC) algorithm
annotates electronic health record (EHR) texts by
leveraging the interface terminology with the highest
coverage, while maintaining optimal time
complexity. In a comprehensive evaluation, we tested
the CFC algorithm on two datasets of varying sizes,
randomly selected from the MIMIC-III database,
using two versions of Cardiology Interface
Terminology (CIT). Compared to the traditional
CFC Annotator: A Cluster-Focused Combination Algorithm for Annotating Electronic Health Records by Referencing Interface
Terminology
203
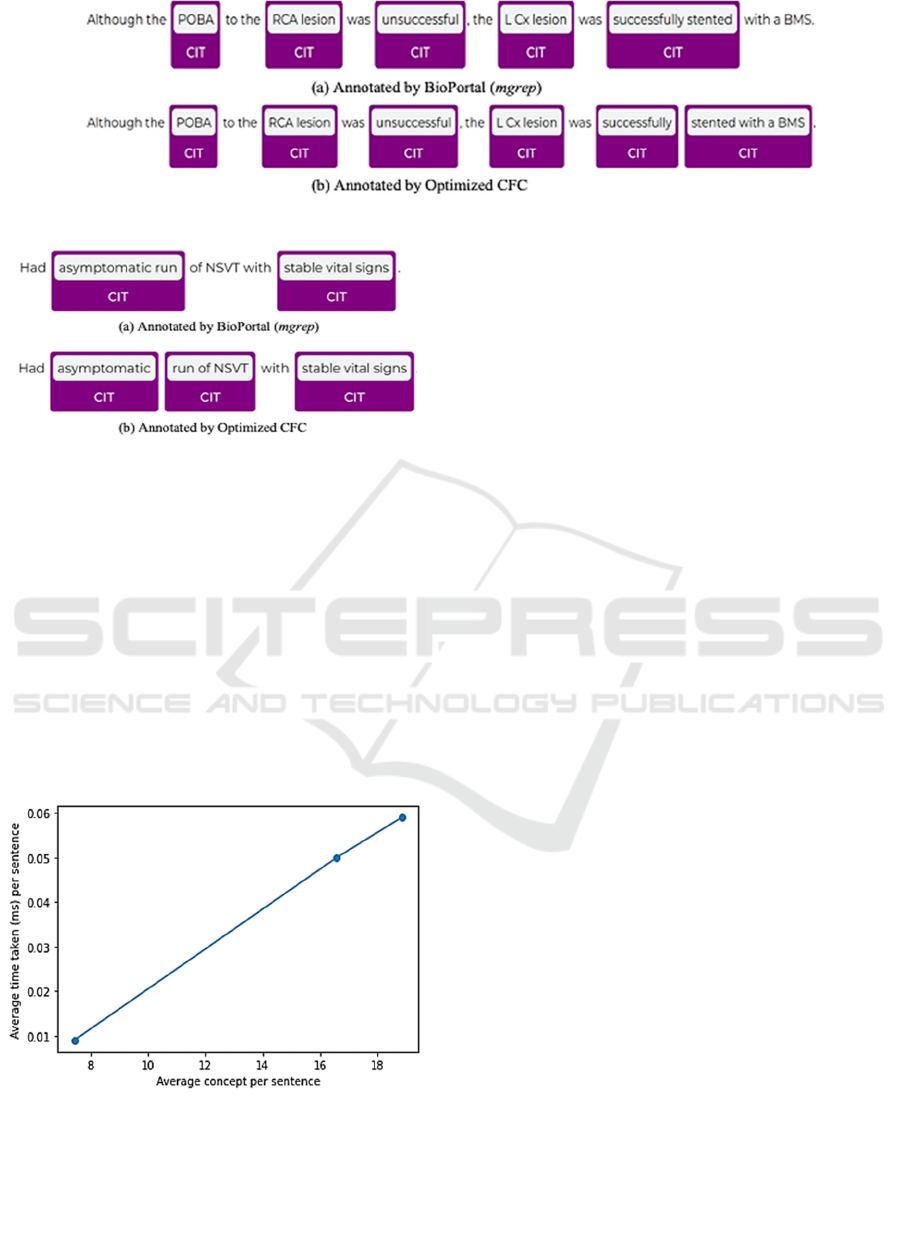
Figure 4: Annotation for Example Two by the BioPortal (mgrep) and optimized CFC annotator.
Figure 5: Annotation for Example Three by the BioPortal
(mgrep) and optimized CFC annotator.
BioPortal (mgrep) longest-chunk annotator, our
algorithm significantly improves the number of
annotated concepts, the number of annotated words,
and overall annotation coverage. Furthermore, the
CFC algorithm offers a substantial reduction in
execution time, making it well-suited for large-scale
datasets and enhancing its scalability and efficiency
in high-volume clinical settings. In Figure 6, we
present the chart of the execution time on different
number of matched concepts per sentence. We
randomly re-generate 150-notes, 500-notes and 1000-
notes for the execution time study.
Figure 6: The execution time on average number of
matched concepts per sentence of three datasets: 150-notes,
500-notes and 1000-notes.
The average number of matched concepts per
sentence for three datasets are: 7.4, 18.9, 16.7
respectively. It is observed that the execution time is
increasing in a linear-like curve. For a sentence with
18 matched concepts, it only takes about 0.06s by our
optimized CFC algorithm. In addition, the tested
1000-notes dataset is processed only in 1.87s which
again demonstrate the power of the algorithm on
annotating enormous scale of dataset.
Despite these advances, the CFC annotator has
certain limitations. It cannot recognize unstructured
phrases that lack direct mappings to the semantics in
the provided terminology. This reliance on the
quality of the interface terminology means that if the
terminology lacks high granularity concepts or fails
to capture specific information, the CFC algorithm
may miss annotating relevant phrases. Therefore,
the success of CFC is closely tied to the
comprehensiveness and detail of the terminology
provided. As such, careful curation and frequent
updates to the interface terminology are essential for
ensuring that annotations remain accurate and
exhaustive in real-world applications.
This performance improvement indicates great
potential for automated annotation on a massive
scale, alleviating the need for labour-intensive
manual annotation. The optimized CFC can thereby
facilitate the creation of high-quality, large-scale
annotated datasets, supporting future training efforts
for Large Language Models and other data-intensive
AI applications.
The optimized CFC annotator’s flexibility also
makes it adaptable for use with other types of
terminologies or ontologies beyond standard
interface terminology. Users can supply their own
terminology and datasets, allowing for customizable
annotations across a wide array of domains.
However, it is important to note that the broader and
more detailed the terminology, the higher the
expected annotation performance. In future work,
we aim to develop related software based on the
optimized CFC algorithm to provide more
interactive annotation services and plan to test it on
larger-scale datasets across diverse domains to
further assess its versatility and robustness.
HEALTHINF 2025 - 18th International Conference on Health Informatics
204

6 CONCLUSIONS
In conclusion, our research introduces a novel
algorithm, the cluster-focused combination algorithm,
designed to overcome the challenges associated with
annotating Electronic Health Record (EHR) text using
interface terminology. This algorithm addresses
critical issues in text annotation by utilizing a dynamic
programming approach, effectively balancing the need
for high annotation coverage and breadth while
mitigating common pitfalls of previous methods. Our
extensive evaluation on benchmark datasets, such as
Mimic III, reveals an improvement in annotation
coverage and captured 5756 missed annotated
concepts by the traditional BioPortal Annotator.
Additionally, the cluster-focused combination
algorithm demonstrates a notable reduction in
execution time by an average of about 8000 times,
enhancing its scalability for large datasets.
These findings make the optimized CFC a highly
effective tool for real-world text annotation tasks that
rely on interface terminology. By providing a more
efficient and comprehensive solution, this work not
only advances the capabilities in EHR text annotation
but also contributes to the broader field of Natural
Language Processing. This is particularly significant
for the development of Large Language Models, which
depend on vast, well-annotated datasets. Our algorithm
paves the way for future innovations in dataset
preparation, promising to streamline and accelerate the
annotation process for large-scale NLP applications.
ACKNOWLEDGMENTS
H. Liu acknowledges startup funds from Montclair
State Univ.
REFERENCES
Aronson, A. R., & Lang, F.-M. (2010). An overview of
MetaMap: Historical perspective and recent advances.
Journal of the American Medical Informatics
Association, 17(3), 229–236.
Blumenthal, D. (2009). Stimulating the adoption of health
information technology. The West Virginia Medical
Journal, 105(3), 28–29.
Bodenreider, O. (2004). The Unified Medical Language
System (UMLS): Integrating biomedical terminology.
Nucleic Acids Research, 32(Database issue), D267–
D270.
Dai, M. (2021). Mgrep. In GitHub repository. GitHub.
https://github.com/daimh/mgrep
Dean, M., Schreiber, A. T., Bechofer, S. K., Harmelen, F.
van, Hendler, J. A., Horrocks, I., MacGuinness, D.,
Patel-Schneider, P. F., & Stein, L. A. (2004).
OWL Web Ontology Language—Reference.
https://api.semanticscholar.org/CorpusID:60998041
Dehkordi, M. K. H. & others. (2023). Using annotation for
computerized support for fast skimming of cardiology
electronic health record notes. 2023 IEEE International
Conference on Bioinformatics and Biomedicine
(BIBM), 4043–4050.
Demner-Fushman, D., Rogers, W. J., & Aronson, A. R.
(2017). MetaMap Lite: An evaluation of a new Java
implementation of MetaMap. Journal of the American
Medical Informatics Association, 24(4), 841–844.
Donnelly, K. (2006). SNOMED-CT: The advanced
terminology and coding system for eHealth. Stud
Health Technol Inform, 121, 279–290.
Jonquet, C., Shah, N. H., & Musen, M. A. (2009). The open
biomedical annotator. Summit on Translational
Bioinformatics, 2009, 56–60.
Kanter, A. S., Wang, A. Y., Masarie, F. E., Naeymi-Rad,
F., & Safran, C. (2008). Interface Terminologies:
Bridging the Gap between Theory and Reality for
Africa. Studies in Health Technology and Informatics,
136, 27–32.
Keloth, V. K., Zhou, S., Lindemann, L., Zheng, L.,
Elhanan, G., Einstein, A. J., Geller, J., & Perl, Y.
(2023). Mining of EHR for interface terminology
concepts for annotating EHRs of COVID patients.
BMC Medical Informatics and Decision Making, 23.
https://api.semanticscholar.org/CorpusID:257106827
Keloth, V., Zhou, S., Einstein, A., Elhanan, G., Chen, Y., &
Geller, et al., J. (2020). Generating Training Data for
Concept-Mining for an ‘Interface Terminology’
Annotating Cardiology EHRs. 2020 IEEE International
Conference on Bioinformatics and Biomedicine
(BIBM).
M, S., & Chacko, A. M. (2020). A Case for Semantic
Annotation Of EHR. 2020 IEEE 44th Annual
Computers, Software, and Applications Conference
(COMPSAC), 1363–1367.
Miles, A., & Bechhofer, S. (2009). SKOS Simple
Knowledge Organization System Reference
.
Musen, M. A., Shah, N. H., Noy, N., Dai, B., Dorf, M.,
Griffith, N., Buntrok, J., Jonquet, C., Montegut, M., &
Rubin, D. (2008). BioPortal: Ontologies and data
resources with the click of a mouse. AMIA ... Annual
Symposium Proceedings. AMIA Symposium, 1223–
1224.
Noy, N. F. & others. (2009). BioPortal: Ontologies and
integrated data resources at the click of a mouse.
Nucleic Acids Research, 37(suppl_2), W170–W173.
Rosenbloom, S. T. & others. (2006). Interface terminologies:
Facilitating direct entry of clinical data into electronic
health record systems. Journal of the American Medical
Informatics Association, 13(3), 277–288.
Rosenbloom, S. T. & others. (2008). A model for evaluating
interface terminologies. Journal of the American
Medical Informatics Association, 15(1), 65–76.
CFC Annotator: A Cluster-Focused Combination Algorithm for Annotating Electronic Health Records by Referencing Interface
Terminology
205

Saeed, M., Lieu, C. C., Raber, G., & Mark, R. G. (2002).
MIMIC II: a massive temporal ICU patient database to
support research in intelligent patient monitoring.
Computers in Cardiology, 641–644.
Savova, G. K. & others. (2010). Mayo clinical Text
Analysis and Knowledge Extraction System
(cTAKES): Architecture, component evaluation and
applications. Journal of the American Medical
Informatics Association, 17(5), 507–513.
HEALTHINF 2025 - 18th International Conference on Health Informatics
206
