
Segmentation of Intraoperative Glioblastoma Hyperspectral Images
Using Self-Supervised U-Net++
Marco Gazzoni
1 a
, Marco La Salvia
1 b
, Emanuele Torti
1 c
, Elisa Marenzi
1 d
, Raquel Leon
2 e
,
Samuel Ortega
3 f
, Beatriz Martinez
2 g
, Himar Fabelo
2,4 h
, Gustavo Callicò
2 i
and
Francesco Leporati
1 j
1
University of Pavia, Department of Electrical, Computer and Biomedical Engineering, Via Ferrata 5, Pavia I-27100, Italy
2
Research Institute for Applied Microelectronics, University of Las Palmas de Gran Canaria (ULPGC), 35017 Las Palmas
de Gran Canaria, Spain
3
Norwegian Institute of Food, Fisheries and Aquaculture Research, 9019 Tromsø, Norway
4
Fundación Canaria Instituto de Investigación Sanitaria de Canarias and the Research Unit, Hospital Universitario de
Gran Canaria Dr. Negrin, Las Palmas de Gran Canaria, Spain
Keywords: Brain Cancer, Computer-Aided Diagnosis, Deep Learning, Disease Diagnosis, Hyperspectral Imaging,
Self-Supervised Learning.
Abstract: Brain tumour resection yields many challenges for neurosurgeons and even though histopathological analysis
can help to complete tumour elimination, it is not feasible due to the extent of time and tissue demand for
margin inspection. This paper presents a novel attention-based self-supervised methodology to improve
current research on medical hyperspectral imaging as a tool for computer-aided diagnosis. We designed a
novel architecture comprising the U-Net++ and the attention mechanism on the spectral domain, trained in a
self-supervised framework to exploit contrastive learning capabilities and overcome dataset size problems
arising in medical scenarios. We operated fifteen hyperspectral images from the publicly available HELICoiD
dataset. Enhanced by extensive data augmentation, transfer-learning and self-supervision, we measured
accuracy, specificity and recall values above 90% in the automatic end-to-end segmentation of intraoperative
glioblastoma hyperspectral images. We evaluated our outcomes with the ground truths produced by the
HELICoiD project, obtaining results that are comparable concerning the gold-standard procedure.
1 INTRODUCTION
Brain and Central Nervous System (CNS) cancers are
ranked in the 12th position in terms of mortality,
concerning both genders, with 248,500 deaths
worldwide in 2022, according to the International
Agency for Research on Cancer (IARC) of the World
a
https://orcid.org/0000-0003-4213-8270
b
https://orcid.org/0000-0003-3724-8213
c
https://orcid.org/0000-0001-8437-8227
d
https://orcid.org/0000-0003-4537-5618
e
https://orcid.org/0000-0002-4287-3200
f
https://orcid.org/0000-0002-7519-954X
g
https://orcid.org/0000-0001-7835-9660
h
https://orcid.org/0000-0002-9794-490X
i
https://orcid.org/0000-0002-3784-5504
j
https://orcid.org/0000-0003-2901-4935
Health Organization (WHO) (Bray et al., 2024).
Particularly, brain tumours represent the most
common CNS cancer type (Bray et al., 2024). The
WHO classifies such tumours into four grades (Louis
et al., 2021) and glioblastoma (GB - Grade 4) is the
deadliest one, with an age-standardized 5-year
Gazzoni, M., La Salvia, M., Torti, E., Marenzi, E., Leon, R., Ortega, S., Martinez, B., Fabelo, H., Callicò, G. and Leporati, F.
Segmentation of Intraoperative Glioblastoma Hyperspectral Images Using Self-Supervised U-Net++.
DOI: 10.5220/0013245900003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 3: VISAPP, pages
633-639
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
633

survival rate in the 2010-2014 period of 4% to 17%
(Girardi et al., 2023).
Currently, the standard treatment for GB tumours
is surgery, followed by radiotherapy or
chemotherapy. To achieve maximal tumour
resection, neurosurgeons use several intraoperative
tools (Fabelo et al., 2016, 2018; Florimbi et al., 2020)
which, nevertheless, exhibit several constraints,
mainly cost and time, and also do not precisely
outline tumour borders (Halicek et al., 2019).
Hyperspectral Imaging (HSI) is a non-invasive,
non-ionizing and label-free technique (Torti et al.,
2023), that is becoming more popular in the context
of cancer detection thanks to recent technological
advances (Kumar et al., 2021; Lu et al., 2014).
Moreover, several studies highlighted that tumour
cells present a unique molecular spectral signature
and reflectance characteristics (Florimbi et al., 2020;
Leon et al., 2021).
During the last decade, Machine and Deep
Learning (ML, DL) solutions emerged as innovative
tools to examine and cluster different cancer types
using HSI (Collins et al., 2021; Jansen-Winkeln et al.,
2021; La Salvia et al., 2023; Salvia et al., 2022).
Concerning intraoperative GB segmentation of HS
images, this research mainly emerged within the
European project HELICoiD (HypErspectraL
Imaging Cancer Detection) (Fabelo et al., 2016).
Here, an in vivo human brain HS database was
created and several ML and basic DL pipelines were
developed (Florimbi et al., 2020). In this field, the
main challenge is retrieving a target ground truth to
supervise ML algorithms, as physicians can only
partially identify the tumour and its boundaries when
performing a diagnosis (Fabelo et al., 2019).
Therefore, HELICoiD-based ML studies comprised
supervised and unsupervised algorithms to overcome
this problem and perform automatic segmentation of
intraoperative-captured HS images.
Lately, self-supervised learning (SSL) is emerging
as a framework to operate small-sized datasets with
limited labelling (Wang et al., 2022; Yue et al., 2022;
Zhu et al., 2022). SSL algorithms work by distilling
representative characteristics from unlabelled and
unstructured data, learning shared and separate
features in a contrastive manner, surpassing supervised
architectures on many domains (Wang et al., 2022).
To the best of the authors' knowledge, no prior
work exists concerning medical brain tumour HS
images and SSL. Hence, here we propose a novel self-
supervised deep learning architecture, an attention-
based U-Net++, as a proof-of-concept to perform the
automatic end-to-end segmentation of fifteen
intraoperative GB HS images retained from the
HELICoiD database (Fabelo et al., 2019).
Figure 1: Taxonomy of intraoperative GB segmentation
ground truth derived by manually correcting HELICoiD
results to smoothen the borders.
2 MATERIALS AND METHODS
2.1 In-Vivo HS Human Brain Dataset
and Pre-Processing
We operated 27 GB HS images derived from the
HELICoiD database (Fabelo et al., 2016; Florimbi et
al., 2020). Data was captured using an intraoperative
HS acquisition system at The University Hospital
Doctor Negrin of Las Palmas de Gran Canaria (Spain)
(Fabelo et al., 2018). The Comité Ético de
Investigación Clínica-Comité de Ética en la
Investigación (CEIC/CEI) of the University Hospital
Doctor Negrin approved the study and the informed
consent was signed by all participating patients.
Physicians labeled each spatial pixel according to
the taxonomy proposed in Fig. 1, employing a semi-
automatic labelling tool based on the Spectral Angle
Mapper (SAM) method (Fabelo et al., 2019). In this
way, a ground-truth map was generated for each HS
image, where the neurosurgeon selected reference
pixels from normal, tumor, hypervascularized and
background classes. Therefore, the SAM algorithm
clustered pixels that resulted similar to the reference
spectral signatures. The tumor tissue class was
assessed by histopathology.
The HELICoiD ML framework starts by pre-
processing the raw HS images captured by the
intraoperative HS acquisition system and ends by
generating a four-color thematic map after
performing supervised and unsupervised
classification (Fig. 2).
The pre-processing chain applied to the HS image
is detailed in (Florimbi et al., 2020), reducing the
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
634
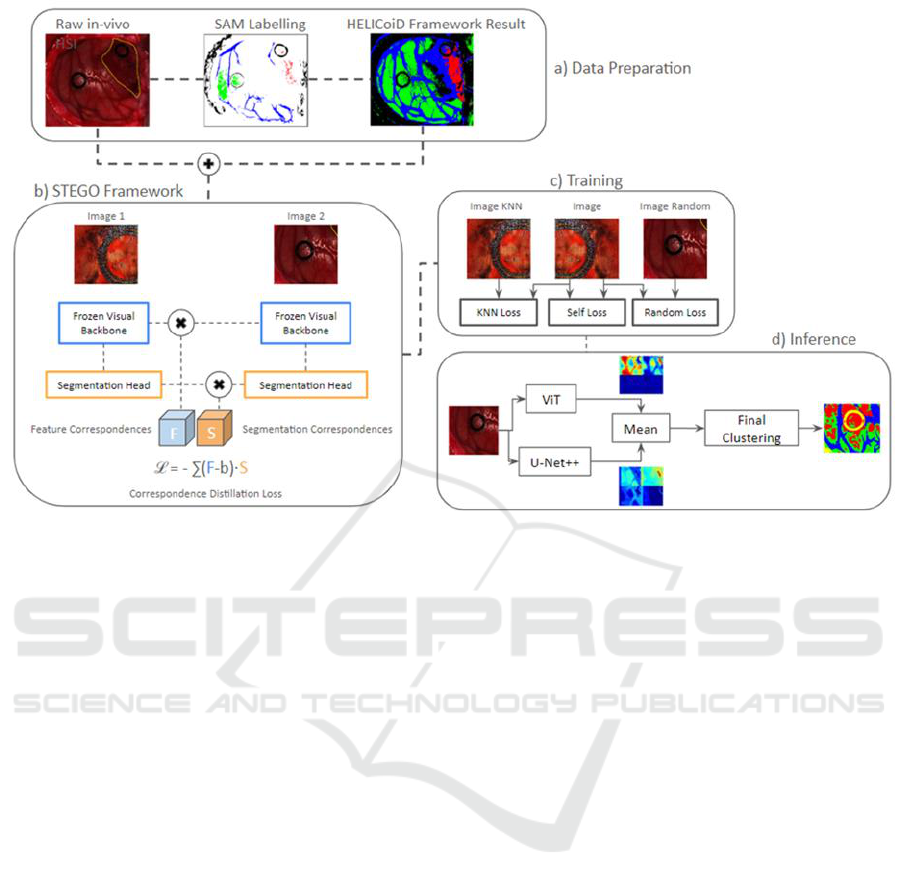
Figure 2: Data preparation, STEGO framework, training and inference resulting from SSL: the raw in-vivo HS images are
pre-processed (Florimbi et al., 2020) and cropped for homogeneity reasons. In addition, in section a), the SAM labelling is
performed only once during the Helicoid framework training to derive the resulting images shown on the right. These, the
pre-processed and the cropped images represent the input to the STEGO framework, in b), employed for segmentation
purposes using supervised learning with U-Net++ backbone network. c) presents the three unsupervised loss functions
adopted in the training phase and how they are combined, while d) details the inference process, using SSL through the
definition of a hybrid network obtained combining the outputs of the parallel execution of U-Net++ and ViT.
instrumentation noise and limiting the curse of data
dimensionality. After preprocessing, HS images
featured 128 spectral bands and cropped to equal
spatial dimensions. The raw HS images were
calibrated for dark noise correction, using a dark and
a white reference (Fabelo et al., 2016; Florimbi et al.,
2020). Moreover, the extreme noisy bands were
removed due to the low performance of the sensor,
obtaining an HS cube of 645 spectral bands. After
that, to avoid redundant information, the HS cube was
decimated to 128 spectral bands and normalized
between 0 and 1.
2.2 Data Partitioning
The dataset employed is composed of 27 images
partitioned at patient-level and divided in 3 subsets:
15 images together with their ground-truths are used
for the supervised training, 11 (without ground-
truths) for the unsupervised training and 1 for test.
Each image has been resized to obtain a spatial
dimension of 198x198; in this way, computational
performance is optimized by evaluating images of the
same dimensions.
2.3 Attention-Based U-Net++,
Self-Supervised STEGO
Framework and Segmentation
Vision Transformer
Here, we propose a novel DL architecture, namely the
attention-based U-Net++, comprising the attention
mechanism along the spectral dimension and the
well-known U-Net++ architecture along the spatial
frame. In recent years, transformer-based
architectures have proven themself worthy of
investigation in vision contexts (Shamshad et al.,
2023).
Following data preparation, we used the attention-
based U-Net++ inside the STEGO (Self-supervised
Transformer with Energy-based Graph
Optimization), a novel framework that distils
unsupervised features into high-quality discrete
semantic labels (Hamilton et al., 2022). We carefully
modified the algorithmic structure, developed from
scratch in MATLAB 2022a (MathWorks, CA, USA),
to receive the GB HS images as input. It extracts the
features from the backbone architecture, the U-Net++
path, and later retains the segmentation results
Segmentation of Intraoperative Glioblastoma Hyperspectral Images Using Self-Supervised U-Net++
635

corresponding to the selected image characteristics
(Fig. 2-b). After that, by adopting a contrastive
learning methodology, the network can learn feature
correspondences in an unsupervised fashion. In fact,
the STEGO core yields a novel contrastive loss
function (Fig. 2-b) designed to encourage features to
form compact clusters while preserving their
relationships across the entire dataset (Hamilton et al.,
2022). In the STEGO framework, the 15 images with
their ground truth have been used for training.
Successively, an unsupervised training has been
performed on the 11 HSI images without ground truth
to derive the loss function. In this phase, the resulting
loss is the summation of the loss output contributions
obtained from the KNN, self and Random loss
functions (Fig. 2-c).
The last step is the inference, where we modified
the U-Net++ standard architecture, designing an
additional parallel path to analyse the spectral
signatures of the HS image after a first pooling step,
set to reduce the networks' parameters (Fig. 2-d).
Hence, we merged the attention-based neural path
and the U-Net++, averaging their outcomes. More
specifically, we implemented a combination of U-
Net++ and a completely trainable vision transformer
(ViT), used to improve the backbone network’s
predictions (Fig. 2-d). Here, the self-supervised
learning to calculate the loss function extracts
features from both the U-Net++ backbone network
(invariant throughout the training phase, allowing to
obtain coherent characteristics even from diverse
images) and the segmentation, represented by the
entire hybrid network (Hamilton et al., 2022). In such
an application, the segmentation part keeps
parameters fixed only in the first step of the training
so that both networks can run in parallel without
affecting the previous training done with tagged
images. In this phase, the 15 ground-truth maps and
their corresponding HS images have been used (as
well as inputs to the STEGO framework).
The networks were trained using a desktop PC
with Windows 10 SO and Intel processor Core i9-
9900X with 3.5 GHz, 128 GB RAM DDR4 with 2667
MHz working frequency, 2 NVIDIA GPUs GeForce
RTX 2080, each with 8 GB of dedicated memory.
Fifteen images were employed for both STEGO
framework and inference, with dimensions of
198×198 pixels × 128 bands, because they showed
both ground-truth for all the classes and were
conformant with the synthetic RGB image. The
HELICoiD framework results were processed by
deleting wrong point classifications to smoothen the
areas. 11 additional HS images with the same
dimensions but without their ground truths have been
used for the unsupervised training, thus leaving one
image to test the entire method.
Since the available data (especially in terms of
pixels) is not sufficient to allow for a purely supervised
approach, data augmentation through the application of
an augmenter function has been done to perform
transformations able to generate additional images. In
particular, spectral noise, global salt and pepper noise,
affine transformations and spectral bands substitutions,
as well as their options management, were applied. In
this way, for each image a series of configurations are
provided together with their occurrence probability,
thus increasing data variability.
Due to the different spatial dimensions between
the images in the database, a sub portion has been
selected as a trade-off between the amount of
information and hardware constraints. In the UNet++
architecture, a series of convolutions is performed:
the first has 128 filters, while successive blocks
employ, respectively, 64, 128, 256, 512 and 1024
convolutions. These are followed by an oversampling
that brings back the original spatial dimensions, with
the number of filters kept constant while their
execution order is inverted, and the last convolutional
layer has the same number of filters as the final
number of classes in the image, that is 4.
The drop out layer has always the same
probability value of 0.5 and the final part of the
network, represented by the combination of softmax
and classification layers.
The first sampling layer of ViT employs an 8×8
window, further changing spatial dimensions to 24×24,
while patches are extracted through a 1×1 window to
select single spectral signatures forcing the network to
focus on the spectral dimension. Such patches are then
projected to have an embedded dimension of 256
(twice the initial number of bands). Four transformer
encoding blocks were used with 8 heads by the
attention layer, whereas the MLP layer projects data
with double their dimensions, hence returning, as
output, the same dimensions of the input. Lastly, all
drop out layers have 0.4 as probability value.
3 RESULTS AND DISCUSSION
We evaluated the outcomes both quantitatively (as
shown in Fig. 3) and qualitatively concerning the
results retrieved from the HELICoiD dataset, since it
represents the safest and most honest way of
performance assessment. Fig. 3 exhibits the set of
evaluation metrics considered in this study.
Qualitatively, concerning the hypervascularized
tissue, the self-supervised architecture proposed in
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
636
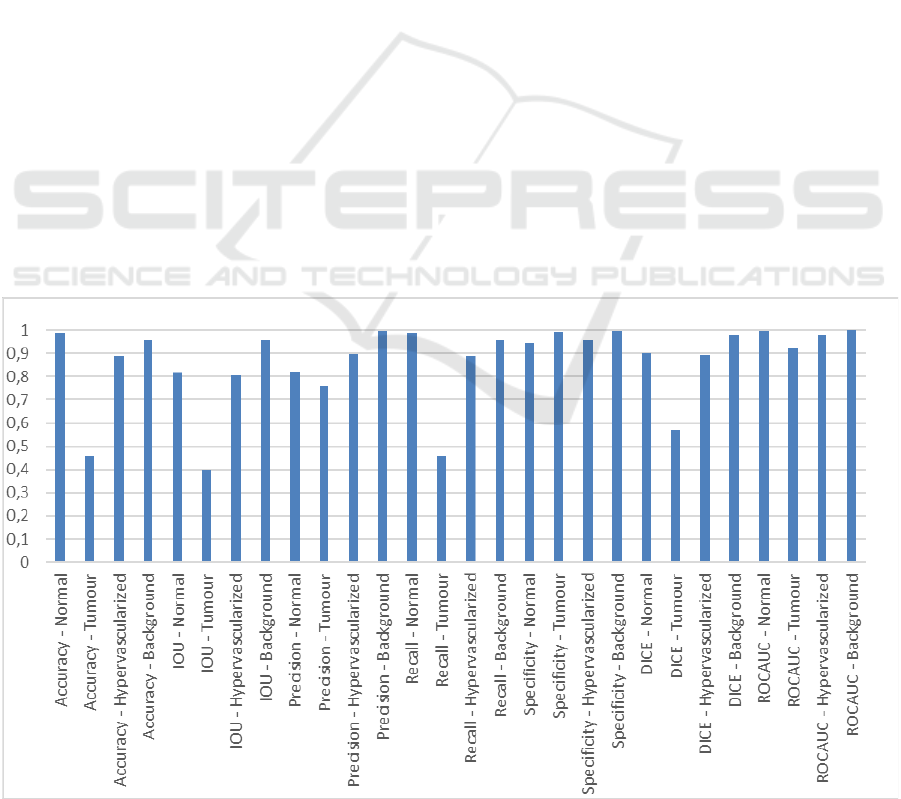
this study can precisely outline this class. In addition,
although the attention-based U-Net++ retains suitable
specificity, recall and accuracy concerning the
tumour class, it yields improvable results for the
normal class. In fact, even though data augmentation
has been performed, the explanation of this
performance is due to the still small number of images
available, resulting in the architecture misclassifying
the background, or wrongly detecting normal
signatures as malignant ones. This issue will be
solved without changing the model architecture when
a bigger dataset is conceived.
Furthermore, we measured competitive inference
times compared to the standard CUDA environment
offered by MATLAB 2022a, hence without a custom
implementation, concerning the HELICoiD
processing times. HELICoiD’s fastest parallel and
best optimized version took 1.68 s to elaborate the
largest image of the database employing a GPU
(Florimbi et al., 2020), whilst our methodology
performs segmentation inference in 6.73 ± 2.58 s,
thoroughly satisfying the real-time constraint
imposed by the intraoperative HS acquisition system
used in the HELICoiD project, which acquired the
images in less than 80 s (Fabelo et al., 2018).
In the case of the hybrid network (combination of
ViT and U-Net++), a first training has been
performed to obtain baseline values, followed by the
self-supervised framework. The first part is divided
into four steps:
1. U-Net++ and ViT pre-training,
2. supervised training, through the adoption of the
STEGO framework, of the single nets for
segmentation purposes,
3. tumours’ segmentation by training of all
networks (through substitution of the last layer
with a weighted classification one to discriminate
the error contribution among all classes of the
entire dataset) and loss function calculus through
unsupervised learning combining the
contributions of KNN, self and random loss
functions,
4. self-supervised training of the attention-based U-
Net++.
Performance of the trained network has been
evaluated on the entire dataset of 27 images through
self-supervised learning and compared to statistical
metrics derived from the confusion matrix (Fig. 4):
accuracy, precision, recall, DICE similarity
coefficient, F1 score, Intersection over Union (IoU)
and ROC’s Area Under Curve (AUC).
4 CONCLUSIONS
In this work we investigated a novel DL methodology
targeting the end-to-end semantic segmentation of
hyperspectral images belonging to the HELICoiD
Figure 3: Hybrid network segmentation results.
Segmentation of Intraoperative Glioblastoma Hyperspectral Images Using Self-Supervised U-Net++
637

Figure 4: Example of the segmentation output for a test
image undergoing self-supervised learning. The tumour is
shown in red, while the background is in yellow.
Hypervascularized tissue is in blue and normal tissue is in
green.
dataset. Namely, we researched a self-supervised
algorithm to train an innovative segmentation
architecture. The proposed methodology allows the
end-to-end segmentation of such images, targeting
real-time processing to be employed during open
craniotomy in surgery.
This innovative approach improves the gold-
standard HELICoiD pipeline and it offers competitive
results in terms of classification. We measured
competitive inference results for the identification of
unhealthy tissue, namely exceeding 90% in
specificity and recall. Nonetheless, the framework
exhibits poor performance when the architecture
classifies normal and background image portions as
tumour.
On the other hand, this is an open research topic
which we aim to improve and clarify in further works.
We believe the proposed SSL methodology could
refine medical HS image segmentation, thus brushing
up state of the art computer-aided diagnostic systems.
A further improvement will be the evaluation of our
approach considering broader datasets, including a
higher number of images, potentially coming from
different brain tumours, thus obtaining a general
diagnostic tool.
The proposed methodology could enhance
medical hyperspectral research overcoming labelling
and dataset size challenges.
ACKNOWLEDGEMENTS
This work was supported in part by the Spanish
Government and European Union (FEDER funds) in
the context of TALENT-HExPERIA project, under
the contract PID2020-116417RB-C42
AEI/10.13039/501100011033.
REFERENCES
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R.
L., Soerjomataram, I., & Jemal, A. (2024). Global
cancer statistics 2022: GLOBOCAN estimates of
incidence and mortality worldwide for 36 cancers in
185 countries. CA: A Cancer Journal for Clinicians,
74(3), 229–263. doi: 10.3322/caac.21834
Collins, T., Maktabi, M., Barberio, M., Bencteux, V.,
Jansen-Winkeln, B., Chalopin, C., Marescaux, J.,
Hostettler, A., Diana, M., & Gockel, I. (2021).
Automatic Recognition of Colon and Esophagogastric
Cancer with Machine Learning and Hyperspectral
Imaging. Diagnostics, 11(10), 1810. doi:
10.3390/diagnostics11101810
Fabelo, H., Ortega, S., Kabwama, S., Callico, G. M.,
Bulters, D., Szolna, A., Pineiro, J. F., & Sarmiento, R.
(2016). HELICoiD project: a new use of hyperspectral
imaging for brain cancer detection in real-time during
neurosurgical operations (D. P. Bannon, Ed.; p.
986002). doi: 10.1117/12.2223075
Fabelo, H., Ortega, S., Lazcano, R., Madroñal, D., M.
Callicó, G., Juárez, E., Salvador, R., Bulters, D.,
Bulstrode, H., Szolna, A., Piñeiro, J., Sosa, C., J.
O’Shanahan, A., Bisshopp, S., Hernández, M., Morera,
J., Ravi, D., Kiran, B., Vega, A., … Sarmiento, R.
(2018). An Intraoperative Visualization System Using
Hyperspectral Imaging to Aid in Brain Tumor
Delineation. Sensors, 18(2), 430. doi:
10.3390/s18020430
Fabelo, H., Ortega, S., Szolna, A., Bulters, D., Pineiro, J.
F., Kabwama, S., J-O’Shanahan, A., Bulstrode, H.,
Bisshopp, S., Kiran, B. R., Ravi, D., Lazcano, R.,
Madronal, D., Sosa, C., Espino, C., Marquez, M., De
La Luz Plaza, M., Camacho, R., Carrera, D., …
Sarmiento, R. (2019). In-Vivo Hyperspectral Human
Brain Image Database for Brain Cancer Detection.
IEEE Access, 7, 39098–39116. doi:
10.1109/ACCESS.2019.2904788
Florimbi, G., Fabelo, H., Torti, E., Ortega, S., Marrero-
Martin, M., Callico, G. M., Danese, G., & Leporati, F.
(2020). Towards Real-Time Computing of
Intraoperative Hyperspectral Imaging for Brain Cancer
Detection Using Multi-GPU Platforms. IEEE Access,
8, 8485–8501. doi: 10.1109/ACCESS.2020.2963939
Girardi, F., Matz, M., Stiller, C., You, H., Marcos Gragera,
R., Valkov, M. Y., Bulliard, J.-L., De, P., Morrison, D.,
Wanner, M., O’Brian, D. K., Saint-Jacques, N.,
Coleman, M. P., Allemani, C., Bouzbid, S., Hamdi-
Chérif, M., Kara, L., Meguenni, K., Regagba, D., …
Lewis, C. (2023). Global survival trends for brain
tumors, by histology: analysis of individual records for
556,237 adults diagnosed in 59 countries during 2000–
2014 (CONCORD-3). Neuro-Oncology, 25(3), 580–
592. doi: 10.1093/neuonc/noac217
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
638

Halicek, M., Fabelo, H., Ortega, S., Callico, G. M., & Fei,
B. (2019). In-Vivo and Ex-Vivo Tissue Analysis
through Hyperspectral Imaging Techniques: Revealing
the Invisible Features of Cancer. Cancers, 11(6), 756.
doi: 10.3390/cancers11060756
Hamilton, M., Zhang, Z., Hariharan, B., Snavely, N., &
Freeman, W. T. (2022). Unsupervised Semantic
Segmentation by Distilling Feature Correspondences.
Jansen-Winkeln, B., Barberio, M., Chalopin, C., Schierle,
K., Diana, M., Köhler, H., Gockel, I., & Maktabi, M.
(2021). Feedforward Artificial Neural Network-Based
Colorectal Cancer Detection Using Hyperspectral
Imaging: A Step towards Automatic Optical Biopsy.
Cancers, 13(5), 967. doi: 10.3390/cancers13050967
Kumar, D., & Kumar, D. (2021). Hyperspectral Image
Classification Using Deep Learning Models: A
Review. Journal of Physics: Conference Series,
1950(1), 012087. doi: 10.1088/1742-
6596/1950/1/012087
La Salvia, M., Torti, E., Gazzoni, M., Marenzi, E., Leon,
R., Ortega, S., Fabelo, H., Callicó, G., & Leporati, F.
(2023). AI-based segmentation of intraoperative
glioblastoma hyperspectral images. N. J. Barnett, A. A.
Gowen, & H. Liang (Eds.), Hyperspectral Imaging and
Applications II (p. 12). SPIE. doi: 10.1117/12.2646782
Leon, R., Fabelo, H., Ortega, S., Piñeiro, J. F., Szolna, A.,
Hernandez, M., Espino, C., O’Shanahan, A. J.,
Carrera, D., Bisshopp, S., Sosa, C., Marquez, M.,
Morera, J., Clavo, B., & Callico, G. M. (2021). VNIR–
NIR hyperspectral imaging fusion targeting
intraoperative brain cancer detection. Scientific
Reports, 11(1), 19696. doi: 10.1038/s41598-021-
99220-0
Louis, D. N., Perry, A., Wesseling, P., Brat, D. J., Cree, I.
A., Figarella-Branger, D., Hawkins, C., Ng, H. K.,
Pfister, S. M., Reifenberger, G., Soffietti, R., von
Deimling, A., & Ellison, D. W. (2021). The 2021
WHO Classification of Tumors of the Central Nervous
System: a summary. Neuro-Oncology, 23(8), 1231–
1251. doi: 10.1093/neuonc/noab106
Lu, G., & Fei, B. (2014). Medical hyperspectral imaging:
a review. Journal of Biomedical Optics, 19(1), 010901.
doi: 10.1117/1.JBO.19.1.010901
Salvia, M. La, Torti, E., Gazzoni, M., Marenzi, E., Leon,
R., Ortega, S., Fabelo, H., Callico, G. M., & Leporati,
F. (2022). Attention-based Skin Cancer Classification
Through Hyperspectral Imaging. 2022 25th Euromicro
Conference on Digital System Design (DSD), 871–
876. doi: 10.1109/DSD57027.2022.00122
Shamshad, F., Khan, S., Zamir, S. W., Khan, M. H., Hayat,
M., Khan, F. S., & Fu, H. (2023). Transformers in
medical imaging: A survey. Medical Image Analysis,
88, 102802. doi: 10.1016/j.media.2023.102802
Torti, E., Gazzoni, M., Marenzi, E., Leon, R., Callicò, G.
M., Danese, G., & Leporati, F. (2023). An Attention-
Based Parallel Algorithm for Hyperspectral Skin
Cancer Classification on Low-Power GPUs. 2023 26th
Euromicro Conference on Digital System Design
(DSD), 111–116. doi:
10.1109/DSD60849.2023.00025
Wang, Y., Albrecht, C. M., Braham, N. A. A., Mou, L., &
Zhu, X. X. (2022). Self-Supervised Learning in
Remote Sensing: A review. IEEE Geoscience and
Remote Sensing Magazine, 10(4), 213–247. doi:
10.1109/MGRS.2022.3198244
Yue, J., Fang, L., Rahmani, H., & Ghamisi, P. (2022). Self-
Supervised Learning With Adaptive Distillation for
Hyperspectral Image Classification. IEEE
Transactions on Geoscience and Remote Sensing, 60,
1–13. doi: 10.1109/TGRS.2021.3057768
Zhu, M., Fan, J., Yang, Q., & Chen, T. (2022). SC-
EADNet: A Self-Supervised Contrastive Efficient
Asymmetric Dilated Network for Hyperspectral Image
Classification. IEEE Transactions on Geoscience and
Remote Sensing, 60, 1–17. doi:
10.1109/TGRS.2021.3131152
Segmentation of Intraoperative Glioblastoma Hyperspectral Images Using Self-Supervised U-Net++
639
