
Investigating the Propagation of CT Acquisition Artifacts
along the Medical Imaging Pipeline
Jakob Peischl and Renata G. Raidou
a
Institute of Visual Computing & Human-Centered Technology, TU Wien, Favoritenstrasse 9-11, 1060, Vienna, Austria
Keywords:
Uncertainty Quantification and Propagation, Medical Imaging, Medical Image Analysis and Visualization.
Abstract:
We propose a framework to support the simulation, exploration, and analysis of uncertainty propagation in
the medical imaging pipeline—exemplified with artifacts arising during CT acquisition. Uncertainty in the
acquired data can affect multiple subsequent stages of the medical imaging pipeline, as artifacts propagate and
accumulate along the latter, influencing the diagnostic power of CT and potentially introducing biases in even-
tual decision-making processes. We designed and developed an interactive visual analytics framework that
simulates real-world CT artifacts using mathematical models, and empowers users to manipulate parameters
and observe their effects on segmentation outcomes. By extracting radiomics features from artifact-affected
segmented images and analyzing them using dimensionality reduction, we uncover distinct patterns related to
individual artifacts or combinations thereof. We demonstrate our proposed framework on use cases simulat-
ing the effects of individual and combined artifacts on segmentation outcomes. Our application supports the
effective and flexible exploration and analysis of the impact of uncertainties on the outcomes of the medical
imaging pipeline. Initial insights into the nature and patterns of the simulated artifacts could also be derived.
1 INTRODUCTION
Medical imaging is foundational to modern medicine,
aiding in disease prevention, diagnosis, and treatment.
Imaging modalities like Computed Tomography (CT)
and Magnetic Resonance Imaging (MRI) provide
patient-specific information based on physical prin-
ciples that capture dedicated tissue characteristics to
support clinical decisions (Gillmann et al., 2021).
However, data uncertainties stemming from several
factors, such as low spatial resolution, artifacts, or
hardware/software issues, are often present (Ristovski
et al., 2014; Gillmann et al., 2021). This causes dis-
crepancies between the physical properties of imaged
tissue and the respective image representation. These
uncertainties are subsequently propagated through
the medical imaging pipeline, affecting all the remain-
ing steps of processing, segmentation, mapping, and
rendering (Preim and Botha, 2013)—potentially, im-
pacting the accuracy of diagnosis and treatment.
Uncertainty in medical imaging can affect mul-
tiple stages of the medical imaging pipeline, with
significant challenges at each step (Schlachter et al.,
2019). For example, in CT data acquisition we often
encounter artifacts due to patient motion and partial
a
https://orcid.org/0000-0003-2468-0664
volume artifacts. Such artifacts may further propa-
gate and accumulate during pre-processing, segmen-
tation, and mapping, influencing the diagnostic power
of the employed data and potentially introducing bi-
ases in eventual decision-making processes (Raidou,
2018). For decisions involving artificial intelligence
(AI), uncertainty can additionally compromise model
reliability (Zhou et al., 2021). Whether the “decision
maker” is an AI model or a human, recognizing and
mitigating uncertainties is essential.
This paper focuses on understanding the propaga-
tion of acquisition uncertainty, exemplified with CT
imaging. CT is integral to numerous clinical pro-
cesses, for which the presence of potential uncertain-
ties is often critical. Motion artifacts, partial vol-
ume effects, and ring artifacts are among the most
prevalent uncertainties that impact CT imaging qual-
ity (Preim and Botha, 2013). By examining these ar-
tifacts, we aim to map the ways uncertainties in the
acquisition step of the medical imaging pipeline af-
fect subsequent steps—and in particular, the last step
of analysis.
To this end, we introduce a methodology to simu-
late and investigate the propagation of CT acquisition
uncertainty using mathematical models that describe
the aforementioned, common artifacts. Our proposed
752
Peischl, J. and Raidou, R. G.
Investigating the Propagation of CT Acquisition Artifacts along the Medical Imaging Pipeline.
DOI: 10.5220/0013254200003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 1: GRAPP, HUCAPP
and IVAPP, pages 752-764
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

methodology involves a user-in-the-loop approach for
constructing uncertainty pipelines by combining ar-
tifacts to model their effects on the acquired im-
ages, and their impact on the subsequent segmenta-
tion. Making use of radiomics feature extraction and
unsupervised learning approaches, we visualize and
analyze segmentation outcomes from artifact-affected
images. In this way, we can evaluate in real time
how acquisition uncertainty has propagated through
the pipeline and the magnitude of its impact on di-
agnostic information. Although our focus is on CT
artifacts, the proposed methodology can be adapted
to other modalities, allowing for broader applicability
in medical imaging uncertainty research.
Our contributions to the field of uncertainty visu-
alization include the introduction of a novel frame-
work for acquisition uncertainty analysis in medical
imaging, an interactive application for the investiga-
tion of simulated uncertainty pipelines, and a demon-
stration of use cases that illustrate the potential of our
approach in a real-life setting.
2 RELATED WORK
In this section, we provide an overview of different
uncertainty types and examine relevant literature on
uncertainty quantification and propagation, with a fo-
cus on medical imaging.
Uncertainty types are classified as either epis-
temic or aleatoric (Potter et al., 2012). Epistemic
uncertainty results from model imperfections and can
be reduced by improved measurements or calibration,
while aleatoric uncertainty arises from inherent ran-
domness and is typically represented with probabil-
ity density functions (PDFs). Given that CT image
artifacts are often reducible or avoidable, our work
focuses on epistemic uncertainty. However, our em-
ployed methods could theoretically extend to aleatoric
uncertainties.
We target specific medical imaging pipeline
steps that we anticipate to be affected by uncer-
tainties (Gillmann et al., 2021). Each step intro-
duces new uncertainties: acquisition uncertainties
may arise from physical model assumptions, segmen-
tation algorithms add model-based and parameter un-
certainties affecting boundary delineations, and map-
ping, e.g., via marching cubes (Lorensen and Cline,
1987), introduces approximations to the derived sur-
face meshes. In this study, segmentation and mapping
are kept invariant to investigate in an isolated manner
uncertainties only from the acquisition step.
(Ristovski et al., 2014) and (Gillmann et al., 2021)
recently authored surveys for uncertainty classifica-
tion in medical applications. Ristovski et al. clas-
sify uncertainties using random fields to represent ar-
bitrary probability distribution functions. By deter-
mining the mathematical characteristics of a random
field, they provide a framework to consider uncer-
tainty propagation behavior. Gillmann et al. review
state-of-the-art uncertainty-aware visualization tech-
niques related to medical imaging applications, ex-
plore pipeline combinations, and identify challenges
in creating comprehensive uncertainty-aware medical
imaging workflows.
Uncertainty quantification and propagation re-
quires formalized mathematical models to track how
uncertainties propagate within an imaging pipeline. A
general formulation utilizes PDFs, where each input
X
i
of the input vector X = (X
1
, . . . , X
n
)
T
is given as a
PDF f
X
i
(x
i
). The challenge is then to find the PDF of
the output Y after it has been transformed by the func-
tion g, i.e., finding f
Y
(y) where Y = g(X). Various ap-
proaches for determining the output uncertainty in the
so-called forward uncertainty quantification problem
exist (Brodlie et al., 2012). For complex uncertain-
ties, Monte-Carlo-based techniques that approximate
f
Y
(y) are often necessary (Brodlie et al., 2012). A
primary drawback of these methods is computational
inefficiency due to slow convergence. Analytical so-
lutions would be more efficient but are unavailable for
arbitrary PDF propagation.
Uncertainty quantification has become a relevant
aspect of a wide range of scientific fields (Zhang,
2021). Uncertainty-handling frameworks have
been proposed for instance by (Roy and Oberkampf,
2011) to model, estimate, and propagate through any
scientific model uncertainties. (Wu et al., 2012) focus
on explorative aspects of uncertainty and build an in-
teractive tool that visualizes uncertainty flows along a
data processing pipeline—under the assumption that
input uncertainties follow a multivariate normal dis-
tribution. Again, the output uncertainty is estimated
using Monte Carlo sampling.
The application of uncertainty quantification
within medical imaging has primarily focused on vi-
sualization and single-stage analysis of uncertainty
within images. For example, (Howard et al., 2014)
use sampling-based CT uncertainty quantification,
and (Gillmann et al., 2017) employ analytical meth-
ods to highlight image noise as a primary uncertainty
source. (Tian and Samei, 2016) model CT quantum
noise, and (Gravel et al., 2004) examine noise pro-
files across modalities. (Diwakar and Kumar, 2018)
consolidate insights into CT noise, highlighting de-
noising as a key challenge.
There is limited literature on uncertainty prop-
agation across multiple stages of medical imaging
Investigating the Propagation of CT Acquisition Artifacts along the Medical Imaging Pipeline
753

Acquisition
Artifacts
. . .
Pipeline Groups
. . .
. . .
. . .
. . .
Segmentation
. . .
. . .
. . .
. . .
Feature
Extraction
f
1
.
.
.
f
m
, . ..
f
1
.
.
.
f
m
, . ..
f
1
.
.
.
f
m
, . ..
Data
Analysis
PCA
and
t-SNE
Figure 1: Overview of the workflow adopted in our approach. The workflow comprises five steps: acquisition, artifact gen-
eration, pipeline grouping, segmentation, and outcome analysis (which consists of two substeps: radiomics feature extraction
and analysis based on dimensionality reduction).
pipelines. A notable exception is Diffusion Ten-
sor Imaging (DTI), where (Behrens et al., 2003)
laid the groundwork for uncertainty propagation from
diffusion-weighted signals to tensor-derived quanti-
ties. Yet, DTI findings are not generalizable to CT
due to modality-specific differences in imaging re-
construction and clinical use cases. Recent advance-
ments in deep learning (DL) have created interest in
both epistemic and aleatoric uncertainty quantifica-
tion and propagation within medical imaging (Feiner
et al., 2023). However, CT artifacts affecting DL
models in real-world scenarios remain underexplored.
Only (Athanasiou et al., 2015) explored multi-stage
error propagation within imaging pipelines for plaque
classification, linking acquisition and segmentation
errors to classification accuracy. In contrast, our work
evaluates outcomes based on radiomics feature com-
parisons to obtain a holistic view of CT uncertainties
across pipeline stages.
3 METHODOLOGY
In this chapter, we present our proposed methodol-
ogy for the interactive quantification and propaga-
tion of CT uncertainties through the medical imaging
pipeline. The workflow involves five stages, which
are depicted in Fig. 1 and further described in the up-
coming subsections. We first generate an uncertainty-
free base image during the image acquisition stage
(Sec. 3.1). Users can then add artifacts, whose effects
are applied to this base image in the artifacts genera-
tion stage (Sec. 3.2). The pipeline grouping stage al-
lows users to combine different artifacts and specify
valid parameters for each of them (Sec. 3.3). We then
proceed with the segmentation step to calculate seg-
mentations of the artifact-affected images (Sec. 3.4).
Next, we extract radiomics features from the outcome
segmentations and comparatively analyze them using
a combination of principal component analysis (PCA)
and t-distributed stochastic neighbor embedding (t-
SNE), inspired by the previous work of (Reiter et al.,
2018) (Sec. 3.5). This is the outcome analysis stage.
Our interface facilitates these steps, targeting medical
imaging researchers and CT imaging professionals in-
terested in uncertainty quantification and propagation.
3.1 Image Acquisition
At the image acquisition stage, we define the extent,
i.e. the physical dimensions, and spatial resolution
of the CT input volume. A CT dataset can be de-
fined mathematically as a function I : R
3
→ R that
maps a three-dimensional spatial position (x, y, z) to
a corresponding voxel value I(x, y, z). CT voxel val-
ues (i.e. radiodensity values) are given in Hounsfield
Units (HU), proportional to the attenuation coefficient
normalized to water (Yucel-Finn et al., 2023). Users
can define voxel values through implicit modeling or
direct image import. In the former case, users employ
constructive solid geometry to model complex objects
from primitive shapes (Laidlaw et al., 1986). This ap-
proach provides structural information for subsequent
stages and it is showcased in Fig. 2 (a). In the lat-
ter case, users can import image data allowing for a
more realistic data analysis (Fig. 2 (b)). When choos-
ing this option, artifact simulation might be limited,
and the image may already suffer from some kind of
uncertainty. For simplicity, our workflow assumes an
idealized, artifact-free CT acquisition.
IVAPP 2025 - 16th International Conference on Information Visualization Theory and Applications
754
(a)
(b)
Figure 2: In the image acquisition stage, the user can
generate data by (a) implicit modeling or by (b) direct
image import (here, a CT angiogram containing a brain
aneurysm (Klacansky, 2017)).
Figure 3: In the artifacts generation stage, the user defines
which artifacts are to be simulated and in which order. Here,
two artifacts are simulated and combined: a motion artifact
(comprising a Gaussian blur and a ring artifact), and salt
and pepper noise. The order in which artifacts are applied
is specified in the bottom-right corner of the interface.
3.2 Artifacts Generation
In this stage, users define which artifacts they would
like to simulate on the data obtained from the first step
(Fig. 3). Rather than simulating CT acquisition, we
assume an artifact-free CT image, to which artifact
effects are added. This ensures a flexible simulation
across scanning technologies by adjusting artifact pa-
rameters. Multiple artifact sets can also be created
and are combined later, in the stage where pipeline
groups are simulated (Sec. 3.3).
Our framework separates potential CT artifacts
into two types: image artifacts and structure artifacts
(Fig. 4). Image artifacts apply to the entire CT volume
based on voxel-specific radiodensity without needing
structural information. For example, a Gaussian noise
artifact adds random noise to each voxel, as deter-
mined by a user-defined Gaussian distribution. Con-
versely, structure artifacts affect specific structures (or
subregions within an image); thus, they need addi-
tional positional information. For instance, a motion
artifact would affect a structure by shifting it and by
applying a Gaussian blur, as shown in Fig. 3. Simulat-
ing this is only possible if the positional information
of the affected structure is known. Both types of arti-
facts are further discussed below.
Artifacts can be applied sequentially or in parallel
on a given dataset. For image artifacts, we can apply
artifacts sequentially, in parallel, or in a (potentially
nested) combination of both approaches, as shown in
the bottom-right corner of the interface in Fig. 3. In
contrast, the effects of structure artifacts are applied
only in parallel because they are inherently localized
to specific structures within the CT image. Apply-
ing them sequentially would not make sense because
each structure artifact alters independently its associ-
ated structure without influencing others.
3.2.1 Image Artifacts
The image artifacts modeled within our approach, to-
gether with their parameters, are summarized in Fig. 4
(a)–(d). Each of these artifacts follows mathematical
descriptions, as formalized in literature and included
in our Appendix. Salt and pepper noise appears
as isolated bright (“salt”) or dark (“pepper”) voxels,
caused by rare electronic errors and is generally mi-
nor in CT imaging (Lu et al., 2018) (Fig. 4 (a)). De-
spite its limited impact, we include it here for comple-
tion. Parameters of our modeling include radioden-
sity and the relative amount of salt and pepper pixels.
Gaussian noise, observed in low-dose CT images, re-
sults from low photon counts and other electronics-
based noise (Boas and Fleischmann, 2012) (Fig. 4
(b)). Though often modeled by a Poisson distribu-
tion, a Gaussian distribution suffices at high photon
counts (Madhura and Babu, 2017). The parameters
of our modeling are the mean and standard deviation
of the distribution. Cupping artifacts, due to beam
hardening, appear as shading towards the center of a
dense object (e.g. a hard bone structure) (Barrett and
Keat, 2004) (Fig. 4 (c)). The shading appears pro-
nounced in images with larger, isotropic objects like
the skull. Its modeling relies on user-defined param-
Investigating the Propagation of CT Acquisition Artifacts along the Medical Imaging Pipeline
755

(a) Salt and pepper noise (b) Gaussian noise (c) Cupping artifact (d) Ring artifact
(e) Metal artifact (f) Windmill artifact (g) Motion artifact
Figure 4: The artifacts that can be generated as part of our approach, exemplified on a simple implicitly modeled data set.
Subfigures (a)–(d) represent image artifacts, while (e)–(g) represent structure artifacts.
eters for center location and shading intensity. Fi-
nally, ring artifacts are caused by detector miscali-
brations, forming rings around the scanning axis, and
are sometimes mistaken for pathologies (Boas and
Fleischmann, 2012) (Fig. 4 (d)). Modeling parame-
ters include the inner radius, width, radiodensity, and
ring center location.
3.2.2 Structure Artifacts
The structure artifacts modeled within our approach,
together with their parameters, are summarized in
Fig. 4 (e)–(g). Each of these artifacts follows math-
ematical descriptions, as formalized in literature and
included in our Appendix. Metal artifacts are com-
mon in CT due to metal objects that cannot be re-
moved during acquisition (Boas and Fleischmann,
2012) (Fig. 4 (e)). These artifacts stem from effects
such as beam hardening, which causes directional
shading and photon scatter, making shading more
prominent. This artifact is modeled with assump-
tions on attenuation and distance effects, using pa-
rameters for attenuation direction and shading length.
Windmill artifacts appear as evenly spaced, bright
streaks around high-attenuation objects (Barrett and
Keat, 2004) (Fig. 4 (f)). They occur only in helical
CT and increase in number with greater pitch. The
model assumes shading decreases with distance, us-
ing parameters for radiodensity, length, and rotation
per slice. Finally, motion artifacts are frequent, of-
ten due to involuntary or internal movement of the pa-
tient, showing as blurring or double edges (Boas and
Fleischmann, 2012) (Fig. 4 (g)). The model captures
these by transforming structure locations and apply-
ing Gaussian blur to regions of motion, using param-
eters for transformation matrix and blur properties.
3.3 Pipeline Grouping
At this stage, final artifact parameters are set for
customizable pipeline groups. Each group contains
combinations of artifact pipelines (serial, in parallel,
or (nested) combinations thereof) based on a base
pipeline, which users adjust with parameter ranges.
The primary purpose of the parameter ranges is to de-
termine a set of valid values for an artifact parameter.
For example, this enables a user to vary the standard
deviation of a Gaussian artifact contained in the base
pipeline in an interval [a, b] and a step size s, result-
ing in n = ⌊
b − a
s
⌋ distinct artifact pipelines, as shown
in Fig. 1. Since multiple parameter ranges may be
defined per pipeline group and artifact, the total num-
ber of valid configurations for a pipeline group cor-
responds to the number of elements in the Cartesian
product of the parameter spans.
IVAPP 2025 - 16th International Conference on Information Visualization Theory and Applications
756

3.4 Segmentation
During the segmentation stage, the user can segment
a structure of interest. As we are particularly inter-
ested in the propagation of acquisition uncertainties
through the pipeline and their effect on the outcomes,
we choose to keep the segmentation filter simple and
its parameters unchanged. Therefore, we employ a
simple thresholding approach where the user only
needs to select an appropriate threshold for the seg-
mentation. An example of the segmentation outcome
of different pipeline groupings is shown in Fig. 1.
This ensures that we can later compare the resulting
feature values meaningfully since all images have un-
dergone the same segmentation—without having to
additionally consider the effects of the segmentation
step itself on the uncertainty outcomes.
3.5 Outcome Analysis
A quantitative comparison of the artifact effects on
the outcome of the pipeline is not feasible using
segmented images alone. To this end, we adopt a
radiomics-based approach, extracting predefined ra-
diomics features from the segmented outcome of each
artifact-impacted pipeline. Radiomics are quantita-
tive metrics derived from medical images and are of-
ten used to characterize tumors (Gillies et al., 2016).
They consist of a series of features that describe the
morphology, texture, and intensity of a segmenta-
tion. We extract 13 3D shape features and 19 first-
order features, chosen for their time complexity of
O(n log n) and their simpler physical interpretations.
Subsequently, to comparatively explore and ana-
lyze relationships between artifact pipelines, we em-
ploy a t-distributed stochastic neighborhood embed-
ding (t-SNE) (van der Maaten and Hinton, 2008)
and principal component analysis (PCA) (Abdi and
Williams, 2010), similar to the prior work of (Re-
iter et al., 2018). Using a Barnes-Hut algorithm for
t-SNE (van der Maaten, 2014) allows for a reduced
time complexity of O(n logn). We standardize the
feature data for equal weighting and apply t-SNE to
the entire dataset across pipeline groups (e.g. mul-
tiple artifacts with multiple parameterizations) while
using PCA within pipeline groups (e.g. one type of
artifact with several parameterizations), as shown in
Fig. 5. The t-SNE results are represented in an in-
teractive 2D scatterplot, with data points color-coded
by pipeline group (Fig. 5 (a)). Selecting a subset of
points in the t-SNE plot triggers a PCA analysis on
this filtered selection. The PCA output is also shown
in a 2D scatterplot, with insights into the PCs and their
significance using a pie and bar chart (Fig. 5 (b)). This
(a)
(b)
Figure 5: In the outcome analysis step, (a) we use a t-SNE
analysis to compare pipeline groups. The left window dis-
plays a scatterplot of t-SNE results for radiomic features
from each artifact-impacted pipeline. Selecting a data point
in this plot reveals the associated pipeline in the right win-
dow and renders the affected data in the center window.
Here, a cluster of points is selected on the t-SNE view us-
ing a lasso brush. (b) Next, we perform PCA analysis on
the lasso-selected points in (a). The left window shows a
scatterplot of the first two PCs. The center and right win-
dows present a pie chart of explained variances for these
PCs with a bar chart of sorted feature coefficients for PC1,
and renderings and pipeline details for the selected data.
Dark green indicates negative feature coefficients.
combination leverages the ability of t-SNE to detect
non-linear relationships across pipelines and the in-
terpretability of PCA within pipelines.
3.6 Implementation
For the implementation of the framework, C++20 was
chosen for its performance capabilities and low-level
control, while the Qt framework is used for devel-
oping a cross-platform graphical user interface. For
medical image processing, the application employs
the Visualization Toolkit (VTK). Data storage utilizes
the HDF5 format with zlib compression to manage
the large memory requirements effectively. For fea-
ture extraction, PyRadiomics is used. The imple-
mentation of our approach is available on our Open-
Source Repository.
Investigating the Propagation of CT Acquisition Artifacts along the Medical Imaging Pipeline
757
4 RESULTS
This section presents the results of our uncertainty
propagation framework applied to two scenarios. We
describe each scenario and analyze the results, fol-
lowed by a cumulative summary and discussion. The
selected scenarios are intended to demonstrate the ca-
pabilities of our proposed solution with examples rep-
resenting both implicitly modeled and imported data.
Each scenario was designed to enable insights rele-
vant to real-world cases, using CT data and artifact
parameters that reflect realistic parameter ranges.
4.1 Scenario 1: Modeled Data
The first example scenario uses implicitly modeled CT
data. We created a CT structure scene that is chal-
lenging for segmentation by placing structures with
similar radiodensities close to a structure of inter-
est. We added both image and structure artifacts (see
Fig. 4), making segmentation more complex and anal-
ysis more meaningful for a real-life scenario. This
setup aims to simulate applications like cardiac and
pelvic organ CT segmentation, where similar radio-
densities and metal artifacts (e.g., from pacemakers or
hip replacements) commonly introduce uncertainties
complicating processing and analysis tasks. Segmen-
tation was performed using a bandpass threshold fil-
ter with a specific radiodensity range. Although more
complex segmentation algorithms could be used, we
consider this out of the scope of this work.
For this first example scenario, we analyzed the ef-
fects of individual artifacts in isolation. Each pipeline
group contains only artifact pipelines with the same
type of artifact. This way, we can explore all artifacts
simultaneously while keeping the complexity of the
example manageable. For each group, artifacts like
metal and windmill distortions (see Fig. 4 (e–f)) have
been applied to relevant structures, while a motion ar-
tifact has been introduced to affect the appearance of
the sphere of interest. By varying specific artifact pa-
rameters within literature-defined ranges, we gener-
ate seven pipeline groups—each targeting a different
artifact. After parameterization, we have a total of
752 individual pipelines to explore and analyze.
We then proceed with extracting the 32 radiomics
features for each case (13 3D shape features and 19
first-order features). Running a Barnes-Hut-based t-
SNE algorithm (van der Maaten, 2014) on the stan-
dardized feature data extracted from the artifact-
affected images results in a clear separation of the
pipeline groups and, therefore, the different artifact
types (Fig. 6). Nearly all data points of individual
pipeline groups lie within clusters near each other
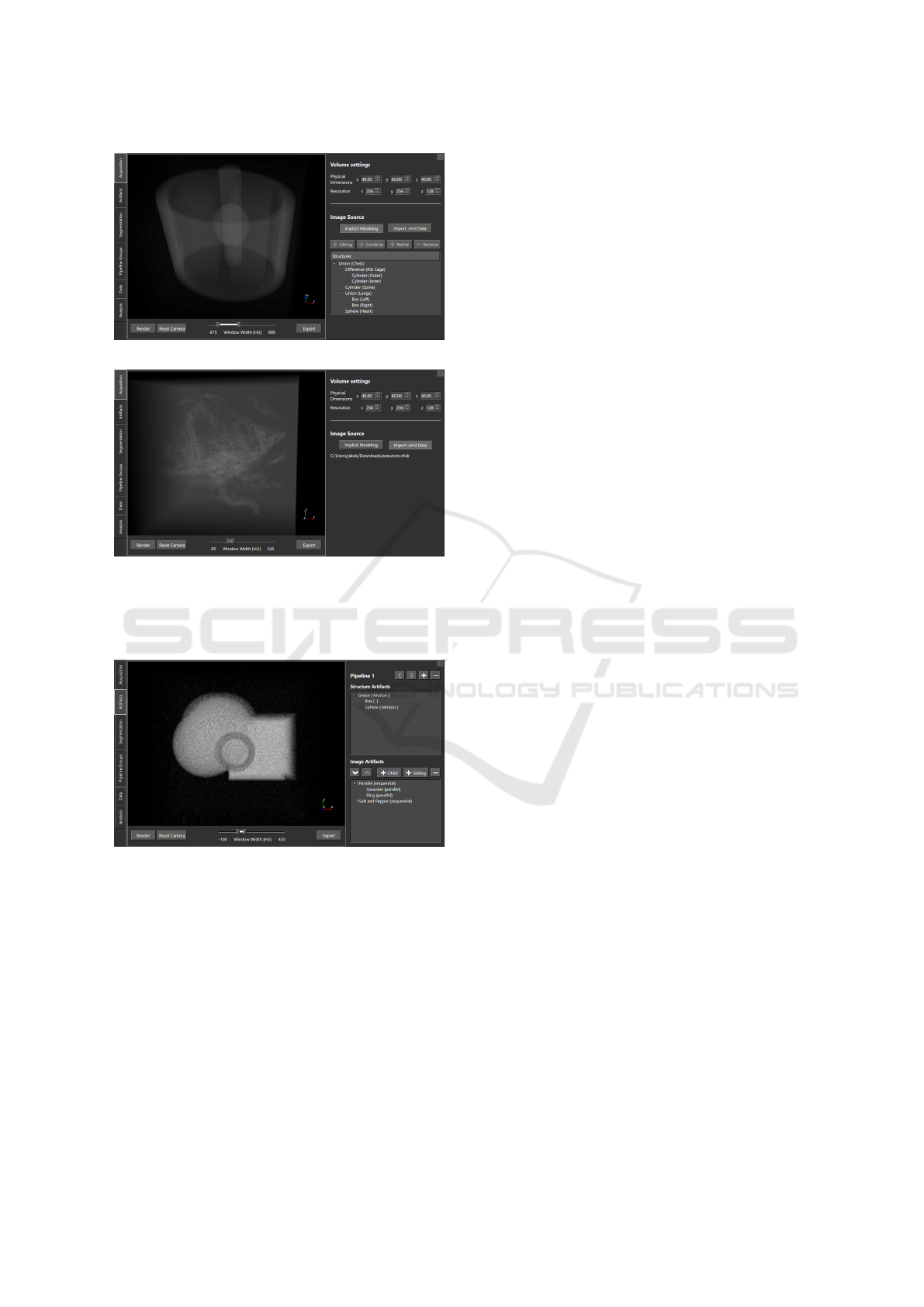
Figure 6: t-SNE for Scenario 1. Small relative distances
suggest similarities between data points. The segmentation
renderings of two pipelines are provided as examples.
Figure 7: Inter-pipeline-group PCAs for Scenario 1: (left)
between Gaussian noise, motion, and metal pipelines;
(right) for windmill artifact pipelines.
while being at a distance from data points correspond-
ing to another artifact type. The outliers result from
extreme parameter values in the artifact generation;
thus, are not discussed any further. Another interest-
ing observation is that for all pipeline group clusters,
the data points are stretched along one main direc-
tion, with one end of the cluster corresponding to the
largest segmentations within the group and the other
end to the smallest ones. Additionally, for the pipeline
groups for which two artifact parameters were varied
(all except for the Gaussian noise), we also noticed the
presence of sub-clusters—potentially indicating com-
peting interests between parameter setups.
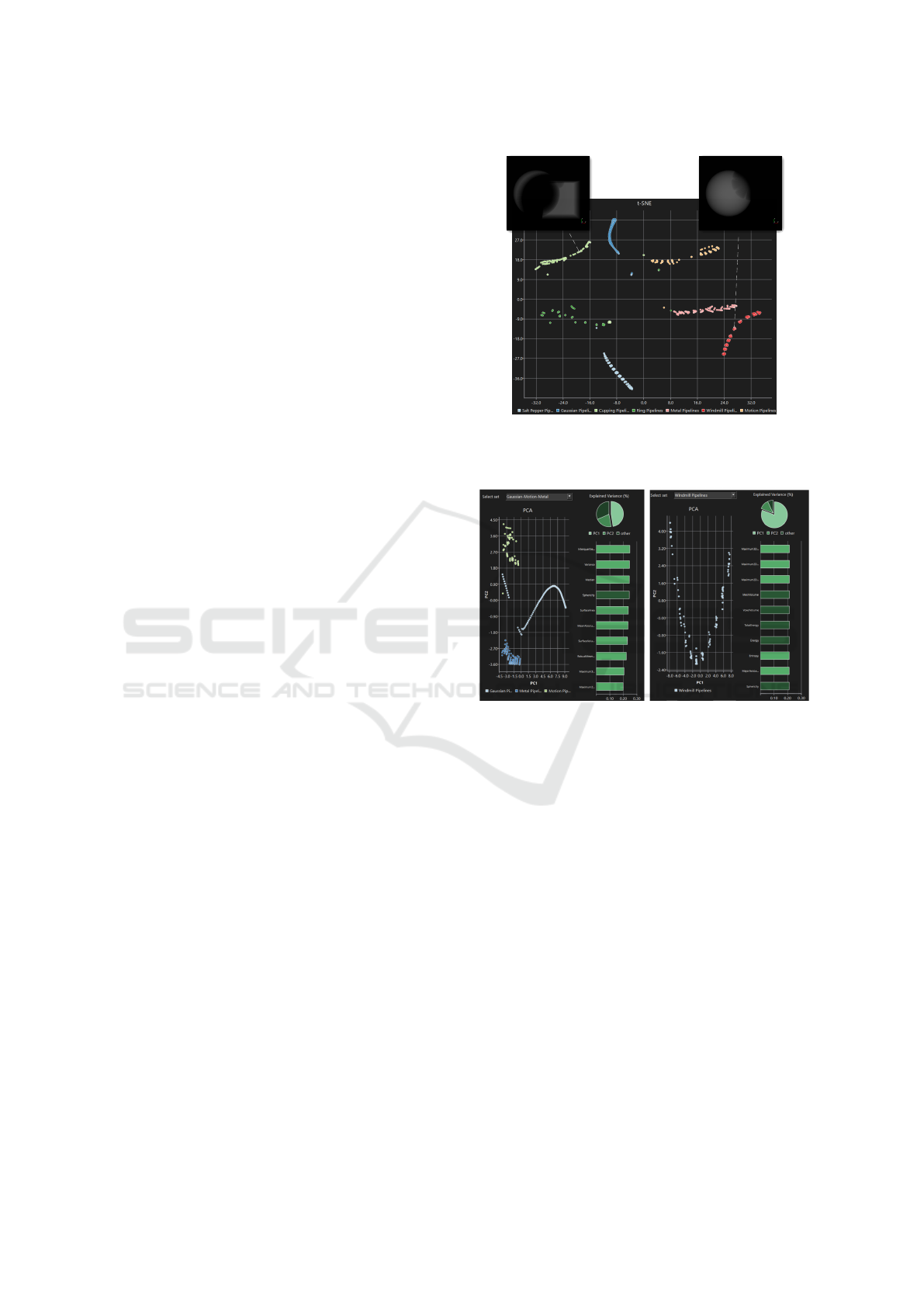
Further analysis with PCA offers insights into
how specific artifact parameters impact segmentation
(Fig. 7). We take a closer look at the PCA for the
windmill pipelines only (Fig. 7, right), as an exam-
ple for deriving insights about the impact of the two
windmill artifact characteristics (i.e. length and angu-
IVAPP 2025 - 16th International Conference on Information Visualization Theory and Applications
758

lar width) on the segmentation. PCA shows the same
sub-clustering pattern observed for the t-SNE, indi-
cating that the method can separate variations of the
two artifact parameters along two dimensions. An-
alyzing the individual data points, we see that the
length increases in the direction of PC1, and the an-
gular width decreases along the direction of PC2.
We examine PC1 more closely since it explains
nearly 81% of the variance in the windmill pipelines
data (see Fig. 7, right; pie chart) and notice that it
assigns equally high priority to many features (see
Fig. 7, right; bar chart). Most (21 out of 32) fea-
tures have an absolute coefficient value within a range
[0.18, 0.21]. This indicates a redundancy between the
employed features. This observation agrees with ex-
pectations of the relationships between individual fea-
tures (Gillies et al., 2016). For instance, the total en-
ergy depends on the energy by a factor of the voxel
volume.
Overall, the shape features dominate the first-
order features (Fig. 7, bar charts left and right). PCA
achieves a precise separation of pipelines by look-
ing at individual features. With increasing length, the
windmill artifact affects larger portions of the sphere
of interest, leading to smaller segmentations. Also,
an increasing length widens the gray-level histogram
of the image due to the gradual nature of the wind-
mill artifact, which explains the high positive weight-
ing for the radiomics feature of entropy. On the other
hand, quantile-related features provide very little in-
formation for PC1 with median or interquartile range
having coefficients close to 0. This is probably due
to the substantial uniformity in the image, given the
assumption that the sphere of interest was modeled as
a homogeneous region.
Performance. Generating the artifact-affected CT im-
ages took the largest time portion (approx. 1 hour 38
minutes). The feature extraction demanded around 16
minutes. The PCA and the t-SNE analysis only took a
negligible amount of time compared to the other two
tasks. Based on the volume settings and implemen-
tation details, the file size of each CT image was de-
termined to be 50.4 MB. For all artifact pipelines, this
amounts to 38 GB, which was reduced to less than 3.9
GB by compression.
4.2 Scenario 2: Real CT Data
In this second scenario, we employ real-world CT
data from a head angiography (Klacansky, 2017),
specifically focusing on the arteries in the right half of
a patient’s head, where an aneurysm is present. The
CT scan, enhanced with a contrast agent, was chosen
due to its relevance in clinical imaging and segmen-
Gaussian
Noise
Artifact
Cupping
Artifact
Ring
Artifact
Salt
Pepper
Noise
Artifact
Salt
Pepper
Noise
Artifact
Gaussian
Noise
Artifact
Cupping
Artifact
Ring
Artifact
Gaussian
Noise
Artifact
Cupping
Artifact
Ring
Artifact
Salt
Pepper
Noise
Artifact
Order A
Order B
Order C
(a)
(b) (c) (d)
Figure 8: (a) Schematic depiction of the order by which the
different artifacts are applied to the dataset in Scenario 2.
(b–d) Visual comparison of order A, B, and C, respectively.
tation tasks, particularly in assessing aneurysm shape
and volume. The detailed and complex arterial struc-
tures make segmentation sensitive to artifacts, and
with only one primary structure of interest, the setup
remains interpretable. This scenario aims to explore
the impact of artifact application order on segmen-
tation outcomes, using four types of image artifacts
(salt and pepper noise, Gaussian noise, cupping, and
ring artifacts). We tested three pipeline configurations
(Fig. 8 (a)):
Order A. This order replicates the order in which ar-
tifacts naturally occur in real CT imaging, with
Gaussian noise and cupping first (as transmission
artifacts), followed by ring (being scanner-based
artifacts) and salt-and-pepper noise (again due to
data transmission) (see Fig. 8 (b)).
Order B. All artifacts are applied in parallel (see
Fig. 8 (c)).
Order C. All artifacts are applied in sequence, with
an arbitrary ordering that is distinct from the other
two for diversity (see Fig. 8 (d)).
In each configuration, artifact parameters re-
mained consistent, though the sequence varied. Vi-
sual analysis of these orders showed slight noise pat-
terns and segmentation differences in fine structures.
To determine an optimal threshold (109 HU) for seg-
mentation, Otsu’s method (Otsu, 1979) was used to
maximize the inter-class variance between the back-
ground and the structure of interest. The same 32 ra-
diomics features were also extracted here for all cases.
The t-SNE analysis displayed five relatively dis-
tinct clusters, although less well-formed than in the
Investigating the Propagation of CT Acquisition Artifacts along the Medical Imaging Pipeline
759

previous scenario (Fig. 9). The algorithm was able
to find similarities within five groups of data points,
albeit they were not as strong as for the first sce-
nario. Each cluster included data points from mul-
tiple pipeline groups, which means that some inter-
pipeline-group similarities exist in our data. This is an
expected result because the pipeline groups use iden-
tical artifact parameters. However, we also see differ-
ences caused by the different orders since Clusters 1,
4, and 5 (Fig. 9) contain unequal shares of data points
from each pipeline group. While Clusters 2 and 3 only
contain data points from Order B and C, respectively.
Salt noise was found consistently across intra-group
points, while other artifact parameters varied across
their full range. Additionally, we find that the posi-
tion of a data point within a cluster no longer reliably
predicts the values of individual artifact parameters.
This suggests that clustering patterns emerged from
combinations of artifact parameters rather than indi-
vidual ones.
PCA was used to interpret the parameter impacts
on segmentation (Fig. 10). PC1 explained over 80%
of the variance in individual groups, primarily due to
the absence of salt noise (Fig. 10, left; pie chart). We
already determined this factor to be influential during
the t-SNE analysis. PCA hardly reveals any informa-
tion about the influence of artifact order because the
separation affects all pipeline groups approximately
the same. Based on the features with the biggest fea-
ture coefficients, we observe that many of our fea-
tures are sensitive to large outliers within the image.
In PC2, which explains only 6.7% of the variance
(Fig. 10, left; pie chart) but is less correlated with
PC1, entropy and uniformity were primary contribu-
tors among the radiomics features. Order C showed
the highest entropy, especially in images with salt
noise, while Order B exhibited the lowest. Order B,
with parallel application, impacts radiodensities less,
and Order A places salt and pepper noise last, reduc-
ing entropy.
Upon looking at the PCA of pipelines with salt
noise (Fig. 10, right), shape features contributed sig-
nificantly to PC1 (Fig. 10, right; pie and bar chart).
Least axis length and minor axis length affect PC1
negatively. Here, these can be interpreted as prefer-
ring a center of mass that aligns better with the center
of the bounding box of a segmentation. This is the
case when the ring artifact affects the region of the
aneurysm strongly because it is the thickest structure
in the image, which can reduce the size of the error
ellipsoid. These quantities stay relatively uniform for
Order B compared to Orders A and C because with
the parallel artifacts of order B, the effects of one ar-
tifact do not influence the effects of the others.
Figure 9: t-SNE for Scenario 2. Five clusters can be re-
trieved, and are highlighted using dashed ellipses and iden-
tified via indices.
Figure 10: PCAs for Scenario 2: (left) PCAs between all
orders (A, B, and C), where artifact pipelines without salt
noise are on the left-hand side and pipelines with salt noise
on the right-hand side; (right) PCAs of artifact pipelines
containing salt noise from all orders.
Performance. The generation of the artifact-affected
CT images took 1 hour 57 minutes, most of which
was used for data compression and writing. On the
other hand, feature extraction took only 37 minutes.
The amount of time PCA and t-SNE needed was also
negligible for this example scenario since we had a
similar number of feature values as in Scenario 1. The
relatively large amount of time spent on compression
and writing was necessary because the images have an
anisotropic resolution of 256 pixels along each axis.
Therefore, the file size of one image was around 100.6
MB, which would have resulted in 76 GB of data but
were compressed to 35.4 GB.
5 DISCUSSION
Flexibility is a strength of our application that is
demonstrated in both use cases. Users are empow-
ered to investigate arbitrary artifact configurations,
IVAPP 2025 - 16th International Conference on Information Visualization Theory and Applications
760

and they may tailor all settings to the imaging tasks of
their interest. As already mentioned in Sec. 2, there is
limited prior work similar to ours. Therefore, our re-
sults are challenging to contextualize within findings
from prior research. However, the fact that the results
of our data analysis agree with literature expectations
or are supported by reasonable explanations is already
a positive indication.
Our approach to evaluating CT artifacts has sev-
eral limitations. While the framework effectively
identifies and quantifies standard artifacts, it may not
adequately capture complex artifacts or interactions
between them. For example, compound effects with
the segmentation step are not currently addressed, as
our approach focuses only on the acquisition uncer-
tainties and their propagation through the pipeline. In
the future, it would be valuable to increase the se-
lection of segmentation algorithms and explore the
impact of this choice (as well as its interplay with
the other steps). Additionally, the extension of this
methodology to other imaging modalities is theoreti-
cally possible. Yet, it presents challenges due to the
unique characteristics and types of artifacts associ-
ated with each modality. Moreover, the actual im-
pact of the findings discussed in Sec. 4 on clinical
decision-making needs a more concrete assessment.
Keeping in mind that the investigated uncertainties
are not the only ones potentially present in an ana-
lytical process (Gillmann et al., 2023; Sacha et al.,
2016), a comprehensive study that includes also the
other types would be necessary. As a future direction,
the framework could integrate a differential analysis
feature to compare selected points in the projected
views with their respective 3D representations. While
the quantitative data and visualizations presented in
the use cases are valuable, evaluating the effective-
ness of our approach in real-world settings with prac-
titioners would be the next step in understanding the
practical implications of our work. For instance, it
would be valuable to evaluate the framework in a set-
ting in which the user learns how uncertainties are
propagated through the medical imaging pipeline.
6 CONCLUSIONS
This work investigated the effects of CT acquisition
artifacts on the subsequent steps and outcomes of the
medical imaging pipeline. Our results indicate that:
• The uncertainty introduced by CT artifacts
impacts segmentation outcomes and feature
extraction—varying by artifact type, order, and
magnitude.
• Our application supports the effective exploration
of the impact of uncertainties on the outcomes of
the medical imaging pipeline.
Our current limitations comprise the choice (and
unexplored effect on the pipeline outcome) of seg-
mentation algorithms and the complexity of artifact
modeling. Addressing these limitations opens inter-
esting directions for future work that are expected to
improve the framework’s generalizability and provide
additional insights into uncertainty propagation.
REFERENCES
Abdi, H. and Williams, L. J. (2010). Principal component
analysis. WIREs Computational Statistics, 2(4):433–
459.
Athanasiou, L. S., Rigas, G., Sakellarios, A., Bouran-
tas, C. V., Stefanou, K., Fotiou, E., Exarchos, T. P.,
Siogkas, P., Naka, K. K., Parodi, O., Vozzi, F., Teng,
Z., Young, V. E., Gillard, J. H., Prati, F., Michalis,
L. K., and Fotiadis, D. I. (2015). Error propagation in
the characterization of atheromatic plaque types based
on imaging. Computer Methods and Programs in
Biomedicine, 121(3):161–174.
Barrett, J. F. and Keat, N. (2004). Artifacts in ct: Recog-
nition and avoidance. RadioGraphics, 24(6):1679–
1691. PMID: 15537976.
Behrens, T., Woolrich, M., Jenkinson, M., Johansen-Berg,
H., Nunes, R., Clare, S., Matthews, P., Brady, M., and
Smith, S. (2003). Characterization and propagation of
uncertainty in diffusion-weighted mr imaging. Mag-
netic resonance in medicine : official journal of the
Society of Magnetic Resonance in Medicine / Society
of Magnetic Resonance in Medicine, 50:1077–88.
Boas, F. E. and Fleischmann, D. (2012). Ct artifacts: Causes
and reduction techniques. Imaging in Medicine,
4:229–240.
Brodlie, K., Osorio, R. S. A., and Lopes, A. (2012). A re-
view of uncertainty in data visualization. In Expand-
ing the Frontiers of Visual Analytics and Visualization,
pages 81–109.
Diwakar, M. and Kumar, M. (2018). A review on ct image
noise and its denoising. Biomedical Signal Processing
and Control, 42:73–88.
Feiner, L. F., Menten, M. J., Hammernik, K., Hager,
P., Huang, W., Rueckert, D., Braren, R. F., and
Kaissis, G. (2023). Propagation and Attribution of
Uncertainty in Medical Imaging Pipelines, page 1–11.
Springer Nature Switzerland.
Gillies, R. J., Kinahan, P. E., and Hricak, H. (2016). Ra-
diomics: images are more than pictures, they are data.
Radiology, 278(2):563–577.
Gillmann, C., Arbel
´
aez, P., Hern
´
andez, J. T., Hagen, H.,
and Wischgoll, T. (2017). Intuitive Error Space Explo-
ration of Medical Image Data in Clinical Daily Rou-
tine. In Kozlikova, B., Schreck, T., and Wischgoll, T.,
editors, EuroVis 2017 - Short Papers. The Eurograph-
ics Association.
Investigating the Propagation of CT Acquisition Artifacts along the Medical Imaging Pipeline
761

Gillmann, C., Maack, R. G. C., Raith, F., P
´
erez, J. F., and
Scheuermann, G. (2023). A taxonomy of uncertainty
events in visual analytics. IEEE Computer Graphics
and Applications, 43(5):62–71.
Gillmann, C., Saur, D., Wischgoll, T., and Scheuermann,
G. (2021). Uncertainty-aware visualization in medi-
cal imaging - a survey. Computer Graphics Forum,
40(3):665–689.
Gravel, P., Beaudoin, G., and de Guise, J. A. (2004). A
method for modeling noise in medical images. IEEE
Transactions on Medical Imaging, 23:1221–1232.
Howard, M., Luttman, A., and Fowler, M. (2014).
Sampling-based uncertainty quantification in decon-
volution of x-ray radiographs. Journal of Computa-
tional and Applied Mathematics, 270:43–51. Fourth
International Conference on Finite Element Methods
in Engineering and Sciences (FEMTEC 2013).
Klacansky, P. (2017). Open scivis datasets.
https://klacansky.com/open-scivis-datasets/.
Laidlaw, D. H., Trumbore, W. B., and Hughes, J. F. (1986).
Constructive solid geometry for polyhedral objects.
In Proceedings of the 13th Annual Conference on
Computer Graphics and Interactive Techniques, SIG-
GRAPH ’86, page 161–170, New York, NY, USA. As-
sociation for Computing Machinery.
Lorensen, W. E. and Cline, H. E. (1987). Marching cubes:
A high resolution 3d surface construction algorithm.
In Proceedings of the 14th Annual Conference on
Computer Graphics and Interactive Techniques, SIG-
GRAPH ’87, page 163–169, New York, NY, USA. As-
sociation for Computing Machinery.
Lu, C.-T., Chen, M.-Y., Shen, J.-H., Wang, L.-L., and Hsu,
C.-C. (2018). Removal of salt-and-pepper noise for
x-ray bio-images using pixel-variation gain factors.
Computers & Electrical Engineering, 71:862–876.
Madhura, J. J. and Babu, D. R. R. (2017). An effective hy-
brid filter for the removal of gaussian-impulsive noise
in computed tomography images. In 2017 Interna-
tional Conference on Advances in Computing, Com-
munications and Informatics (ICACCI), pages 1815–
1820.
Otsu, N. (1979). A threshold selection method from gray-
level histograms. IEEE Transactions on Systems,
Man, and Cybernetics, 9(1):62–66.
Potter, K., Rosen, P., and Johnson, C. R. (2012). From quan-
tification to visualization: A taxonomy of uncertainty
visualization approaches. In Dienstfrey, A. M. and
Boisvert, R. F., editors, Uncertainty Quantification in
Scientific Computing, pages 226–249, Berlin, Heidel-
berg. Springer Berlin Heidelberg.
Preim, B. and Botha, C. P. (2013). Visual Computing
for Medicine: Theory, Algorithms, and Applications.
Morgan Kaufmann Publishers Inc., San Francisco,
CA, USA, 2 edition.
Raidou, R. G. (2018). Uncertainty visualization: Recent
developments and future challenges in prostate can-
cer radiotherapy planning. In EuroRV3 2018: EuroVis
Workshop on Reproducibility, Verification, and Vali-
dation in Visualization, pages 13–17. The Eurograph-
ics Association.
Reiter, O., Breeuwer, M., Gr
¨
oller, E., and Raidou, R.
(2018). Comparative visual analysis of pelvic organ
segmentations. pages 037–041.
Ristovski, G., Preusser, T., Hahn, H. K., and Linsen, L.
(2014). Uncertainty in medical visualization: Towards
a taxonomy. Computers & Graphics, 39:60–73.
Roy, C. J. and Oberkampf, W. L. (2011). A comprehen-
sive framework for verification, validation, and uncer-
tainty quantification in scientific computing. Com-
puter Methods in Applied Mechanics and Engineer-
ing, 200(25):2131–2144.
Sacha, D., Senaratne, H., Kwon, B. C., Ellis, G., and Keim,
D. A. (2016). The role of uncertainty, awareness, and
trust in visual analytics. IEEE Transactions on Visu-
alization and Computer Graphics, 22(1):240–249.
Schlachter, M., Raidou, R. G., Muren, L. P., Preim, B.,
Putora, P. M., and B
¨
uhler, K. (2019). State-of-the-
art report: Visual computing in radiation therapy plan-
ning. In Computer Graphics Forum, volume 38, pages
753–779. Wiley Online Library.
Tian, X. and Samei, E. (2016). Accurate assessment and
prediction of noise in clinical ct images. Medical
Physics, 43(1):475–482.
van der Maaten, L. (2014). Accelerating t-sne using tree-
based algorithms. Journal of Machine Learning Re-
search, 15(93):3221–3245.
van der Maaten, L. and Hinton, G. (2008). Visualizing data
using t-sne. Journal of Machine Learning Research,
9(86):2579–2605.
Wu, Y., Yuan, G.-X., and Ma, K.-L. (2012). Visualiz-
ing flow of uncertainty through analytical processes.
IEEE Transactions on Visualization and Computer
Graphics, 18:2526–2535.
Yucel-Finn, A., Mckiddie, F., Prescott, S., and Griffiths, R.
(2023). A Brief History of Time: From the Big Bang
to Black Holes. Elsevier, 1 edition.
Zhang, J. (2021). Modern monte carlo methods for efficient
uncertainty quantification and propagation: A survey.
WIREs Computational Statistics, 13(5):e1539.
Zhou, S. K., Greenspan, H., Davatzikos, C., Duncan, J. S.,
Van Ginneken, B., Madabhushi, A., Prince, J. L.,
Rueckert, D., and Summers, R. M. (2021). A re-
view of deep learning in medical imaging: Imaging
traits, technology trends, case studies with progress
highlights, and future promises. Proceedings of the
IEEE, 109(5):820–838.
APPENDIX
The symbols x =
h
x
y
z
i
and x
2
= [
x
y
] denote the image
position in all artifact models described below.
Salt and Pepper Noise Artifact
For the salt and pepper noise artifact, we use the fol-
lowing four parameters:
IVAPP 2025 - 16th International Conference on Information Visualization Theory and Applications
762

i
s
radiodensity of salt-pixels
i
p
radiodensity of pepper-pixels
p
s
relative amount of salt-pixels
p
p
relative amount of pepper-pixels
where p
s
+ p
p
≤ 1.0. The output for all values of the
image is then defined as
I
out
(x, Ω) =
i
s
if Ω = 1
i
p
if Ω = 2
I
in
(x) if Ω = 3
(1)
with Ω ∼ Categorical
p
s
, p
p
, 1.0 − (p
s
+ p
p
)
.
Gaussian Noise Artifact
The parameters of Gaussian noise artifacts, following
a Poisson distribution, are:
µ mean of Gaussian distribution
σ standard deviation of Gaussian distribution
for the output image
I
out
(x, T ) = I
in
(x) + T (2)
with T ∼ N (µ, σ
2
).
Cupping Artifact
Our cupping artifacts model uses the following pa-
rameters:
k
I
min
minimum relative radiodensity at center
c =
c
x
c
y
center voxel with maximum shading
Further, we assume that the shading effect increases
linearly towards the center of the artifact to arrive at
the model
I
out
(x) = I
in
(x)
k
I
min
+ (1.0 − k
I
min
)
d
max
− d
c
(x
2
)
d
max
(3)
where d
c
(x
2
) =
|
x
2
− c
|
is the distance from the cen-
ter, and d
max
= max
x
2
∈A
d
c
(x
2
) is the maximum dis-
tance from the center to any voxel in the image.
Ring Artifact
The ring artifacts model takes the following parame-
ters:
r inner radius of the ring
w width of the ring
k
I
relative radiodensity of the ring voxels
c =
c
x
c
y
ring center
The output radiodensities are determined by:
I
out
(x) =
(
k
I
I
in
(x) if
|
x
2
− c
|
∈ [r, r + w]
I
in
(x) else
(4)
Metal Artifact
Metal artifacts modeling makes two central assump-
tions: (a) There is only one direction of highest atten-
uation and one direction of lowest attenuation, which
is perpendicular to the former, and (b) the shading ef-
fect decreases linearly with the distance when moving
away from the boundaries of the structure on a certain
slice. It takes the following parameters:
k
I
min
minimum relative radiodensity adjacent to the
structure in direction of maximum attenuation
l length of effects beginning at structure border
u =
u
x
u
y
direction of highest attenuation
Before we state our model, let the closest point on an
implicit surface S(x) to a given point on the z-value of
the point be
P
S
(x) = argmin
Q∈V
S,z
|
x − Q
|
(5)
with V
S,z
= {x ∈ V | w = z ∧ S(x) = 0} being the set of
points on the curve defined by the cross section of the
xy-plane with the structure S at the current z-value.
Then, we determine the output image using
I
out
(x, S
ξ
) =
(
I
in
(x) if ∄P
S
ξ
(x)
I
in
(x) + R
max,ξ
a
dis
a
dir
else
(6)
where ξ is the implicit structure to which the artifact is
applied, R
max,ξ
is the maximum radiodensity observed
within the structure ξ,
a
dis
= max
0.0, 1.0 −
|
δ
|
l
(7)
is the distance weighting factor with δ = x − P
S
ξ
, and
a
dir
= k
I
max
+
1.0 − 2 (1.0− k
I
max
) cos
2
δ · u
|
δ
|
(8)
is the direction weighting factor.
Windmill Artifact
Windmill artifacts are modeled with the following pa-
rameters:
k
I
max
maximum relative radiodensity adjacent
to the structure on a bright streak
l length of effects beginning at structure border
γ angular width of a single bright streak
φ rotation per slice
The output image is defined by:
I
out
(x, S
ξ
) =
(
I
in
(x) if ∄P
S
ξ
(x)
I
in
(x) + R
max,ξ
k
I
max
a
dis
a
dir
else
(9)
where ξ, R
max,ξ
, a
dis
as well as δ conform to the defi-
nitions provided previously for the metal artifact and
a
dir
= cos
2
π
γ
atan2(δ
y
, δ
x
) + φ (z − z
min
) +
3
4
π
(10)
is the direction weighting factor.
Investigating the Propagation of CT Acquisition Artifacts along the Medical Imaging Pipeline
763

Motion Artifact
Motion artifacts models take the following parame-
ters:
k
I
max
relative radiodensity at the difference region
A linear transformation matrix
r
g
Gaussian blur kernel radius
σ Gaussian blur kernel standard deviation
The output image is defined by:
I
out
(x, S
ξ
) =
(
Γ
r
g
,σ
∗ I
t
(x) if x ∈ V
S
ξ
,A
I
in
(x) else
(11)
where ξ conforms to the definition provided previ-
ously,
V
S
ξ
,A
(x) =
n
x ∈ V
S
ξ
(x) > 0 ∧ S
ξ
A
−1
x
≤ 0
o
(12)
is the set of points in the transformation region,
I
t
(x) = k
I
max
I
in
A
−1
x
(13)
the intensity after transformation, but before blurring,
and Γ
r
g
,σ
is the Gaussian kernel used for blurring.
IVAPP 2025 - 16th International Conference on Information Visualization Theory and Applications
764
