
Czech Salivary Gland Database
Vojt
ˇ
ech Jel
´
ınek
1 a
, Petr Br
˚
uha
1 b
, David Kalfe
ˇ
rt
2 c
and Pavel Nov
´
y
1 d
1
Department of Computer Science and Engineering, Faculty of Applied Sciences,
University of West Bohemia, Univerzitn
´
ı 8, Pilsen, Czech Republic
2
Department of Otorhinolaryngology and Head and Neck Surgery, 1st Faculty of Medicine, University Hospital Motol,
Charles University, V Uvalu 84, Prague 150 06, Czech Republic
vjelinek@students.zcu.cz, {pbruha, novyp}@kiv.zcu.cz, david.kalfert@fnmotol.cz
Keywords:
Salivary Gland Cancer, Clinical Database, Patient Data Management, Data Visualization, Kaplan-Meier
Analysis.
Abstract:
Salivary gland tumors require comprehensive data collection and analysis to support clinical decision-making,
yet existing databases need more focus on specific tumor-related data and visualization tools. This absence
hinders oncologists’ ability to track patient outcomes effectively and identify potential prognostic indicators.
To address this, we developed the Czech Salivary Gland Database (CSGDB), a specialized clinical application
designed to manage patient data and provide visual analytics. The database includes secure and anonymized
data handling alongside Kaplan-Meier survival analysis for outcome visualization. Deployed at the University
Hospital in Motol, CSGDB empowers healthcare professionals with enhanced tools for tracking and analyzing
patient progress, ultimately contributing valuable data and insights to the field of head and neck oncology.
1 INTRODUCTION
Head and neck oncology represents a challenging
field within medical research and clinical practice,
given the diverse types of tumors, unique anatomi-
cal features, and complex treatment paths involved.
Among these, salivary gland tumors are relatively
rare yet require specialized care due to their hetero-
geneous nature, potential malignancy, and impact on
critical physiological functions. Currently, effective
diagnosis, treatment, and monitoring of salivary gland
tumors rely on a combination of clinical expertise
and robust, accurate data. However, existing med-
ical databases and general-purpose electronic health
record (EHR) systems must be better suited for man-
aging the detailed, specific data required for this pa-
tient population.
The challenges associated with these tumors in-
clude tracking long-term outcomes, analyzing treat-
ment effectiveness, and identifying potential prognos-
tic markers—tasks that demand specialized databases
with functionalities tailored to salivary gland oncol-
ogy. Conventional data systems, such as general can-
a
https://orcid.org/0009-0009-8021-4204
b
https://orcid.org/0000-0003-4031-8243
c
https://orcid.org/0000-0003-4369-4113
d
https://orcid.org/0000-0001-9945-1351
cer registries, often lack fields for the specific parame-
ters relevant to these tumors, limiting their usefulness
for clinicians and researchers focused on this area.
Furthermore, these databases frequently do not incor-
porate tools for survival analysis, data visualization,
or detailed cohort tracking, which are essential for un-
derstanding disease progression and informing treat-
ment decisions.
To address these limitations, we developed the
Czech Salivary Gland Database (CSGDB) (Kalfe
ˇ
rt
et al., 2024), a clinical application to support storing,
managing, and analysis of patient data specific to sali-
vary gland tumors. CSGDB provides a secure, spe-
cialized platform for capturing detailed clinical and
demographic data, tracking treatment outcomes, and
visualizing survival trends through integrated Kaplan-
Meier analysis. The design of CSGDB not only ad-
dresses the data storage needs but also emphasizes
user accessibility, ensuring that clinicians can effi-
ciently utilize the system within their workflows.
This paper presents an overview of CSGDB’s de-
sign, architecture, and functionalities. We also dis-
cuss the implementation and deployment of the sys-
tem at the University Hospital in Motol, highlight-
ing its impact on clinical workflow and its potential
contributions to oncology research. CSGDB seeks to
enhance patient care and research capabilities by of-
fering a focused, clinically relevant database, laying
Jelínek, V., Br ˚uha, P., Kalfe
ˇ
rt, D. and Nový, P.
Czech Salivary Gland Database.
DOI: 10.5220/0013254400003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 705-711
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
705
a foundation for future studies and improved clinical
outcomes in head and neck oncology.
2 STATE OF THE ART
As described in the introduction, no specialized head
and neck cancer database exists in the Czech Repub-
lic. From our point of view, it should be a comprehen-
sive database focused on head and neck cancers, col-
lecting complete clinicopathological data and subse-
quently used both for scientific purposes and in every-
day practice. Managing a local database reflects the
conditions and possibilities at that particular work-
place (region, state); thus, the collected data’s value
should be higher. In addition to collecting and or-
ganizing data, such a database should contain tools
for analyzing the various parameters being monitored.
Other features include the ability to update the data
for the monitored patients. Currently, most databases
are based more on retrospective data collection, where
patient data is entered on a specific date (most of-
ten the date the patient is entered into the database)
without subsequent updates. Another advantage of a
local database may be that patients with, e.g., a spe-
cific histopathological type of tumor can be easily re-
trieved, followed by tracing the actual tumor tissue
in the pathology archives for further, e.g., molecular
analyses.
The absence of such a database in the Czech Re-
public is not only on a national scale but also on the
level of local databases of individual clinical depart-
ments.
The national database for head and neck can-
cer established by The Danish Head and Neck Can-
cer Study Group, DAHANCA, see (Overgaard et al.,
2016), which is unique in the world and is designed
for all head and neck cancers, is the inspiration for the
creation of a local CSGDB with aspirations for future
expansion. Its development from a local to a national
database is also inspiring.
In addition to the DAHANCA database, other
more general databases also contain datasets of head
and neck cancers. Two examples from several such
databases are:
• The International Collaboration on Cancer Re-
porting (ICCR) produces common, internation-
ally validated and evidence-based pathology
datasets for cancer reporting for use through-
out the world through broad collaboration be-
tween Pathology Colleges, Societies and major
cancer organizations internationally, spec. Head
and Neck, see (Thompson et al., 2024), (Seethala
et al., 2018);
• The Surveillance, Epidemiology, and End Results
(SEER) Program of the National Cancer Institute
(NCI), https://seer.cancer.gov/; see (Hankey et al.,
1999), (Friedman and Negoita, 2024).
Common to these databases is that most are pri-
marily focused on histopathological parameters of tu-
mours and are therefore more intended for patholo-
gists, i.e. complete clinicopathological data are miss-
ing.
Since we are currently dealing with large salivary
gland tumors, we designed and implemented a CS-
GDB, which is being created within a clinical depart-
ment with data collection from other clinical depart-
ments using a web form (https://slinnezlazy.cz/).
For an idea of the population and incidence
of head and neck cancers in relation to the cited
DANANCA database as an inspirational model, we
present statistical data for the Czech Republic and
Denmark for the period 2022; the data source is the
International Agency for Research on Cancer, see
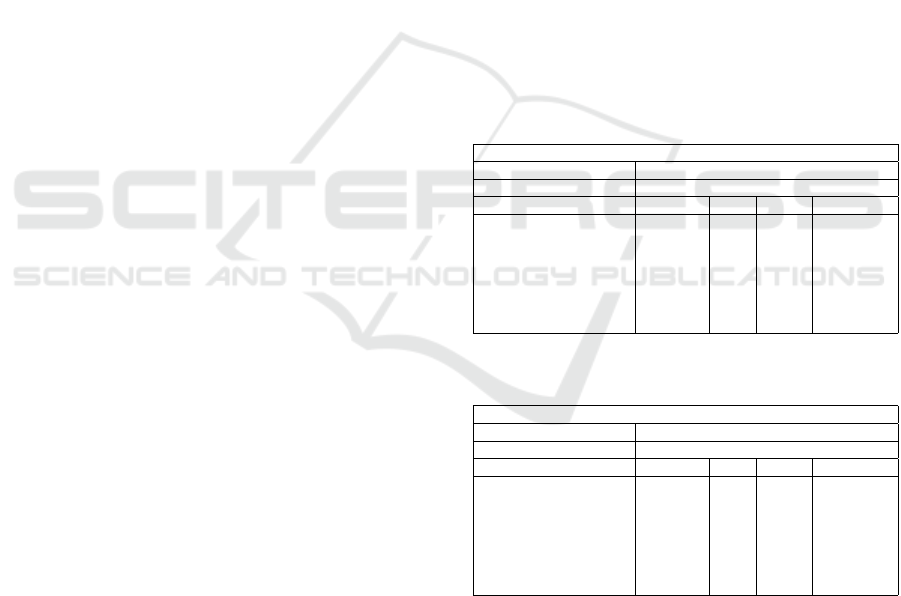
(Ferlay et al., 2024), (Bray et al., 2024), Table 1.
Table 1: Overview table of newly diagnosed head and neck
cancers in the Czech Republic and Denmark in 2022.
CZECHIA - 2022
Population 10 736 782
Number of new Cancer 65 676
Cancer Number (%) Rank Cum. risk
Lip, oral, cavity 955 1.45 15 0.54
Oropharynx 582 0.89 21 0.36
Larynx 465 0.71 23 0.28
Salivary glands 151 0.23 27 0.07
Hypopharynx 150 0.23 28 0.10
Nasopharynx 51 0.08 32 0.03
Head & Neck 2 354 3.58
Available from: https://gco.iarc.who.int/media/globocan/
factsheets/populations/203-czechia-fact-sheet.pdf
DENMARK - 2022
Population 5 834 952
Number of new Cancer 48 840
Cancer Number (%) Rank Cum. risk
Lip, oral, cavity 449 0.92 19 0.47
Oropharynx 450 0.92 18 0.52
Larynx 235 0.48 23 0.25
Salivary glands 82 0.17 28 0.08
Hypopharynx 120 0.25 27 0.13
Nasopharynx 21 0.04 32 0.03
Head & Neck 1 357 2.78
Available from: https://gco.iarc.who.int/media/globocan/
factsheets/populations/208-denmark-fact-sheet.pdf
HEALTHINF 2025 - 18th International Conference on Health Informatics
706
Front-End
Kaplan-Meier Filter
Kaplan-Meier Chart
Patient
Form
Study
Form
Common
Form
Parts
Patients List
Component
Studies List
Component
Add Patient
Component
Add Study
Component
Kaplan-Meier
Component
Renderer Process Main Process
IPC
Back-End
Database
Encryption
Password
Manager
Patients
Manager
Studies
Manager
Basic DB
Operations
API call
API response
function call
function result
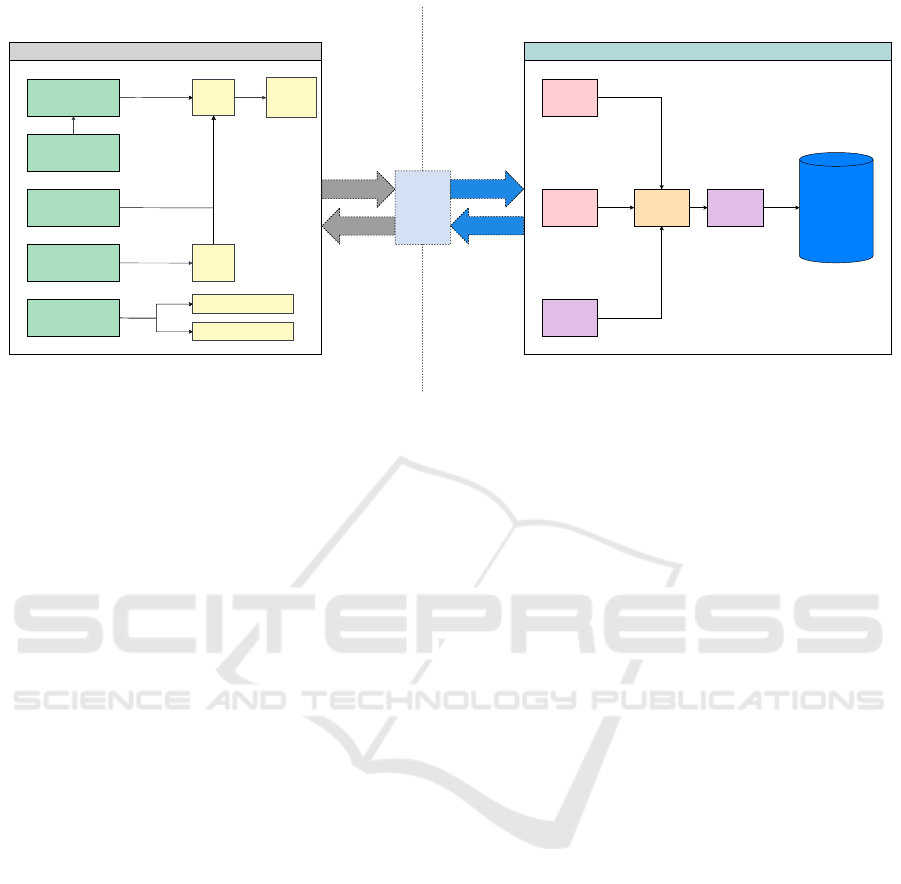
Figure 1: High-level Architecture of the Czech Salivary Gland Database (CSGDB).
3 SYSTEM DESIGN AND
ARCHITECTURE
3.1 Overall Design Goals
The Czech Salivary Gland Database (CSGDB) design
focuses on three main objectives: clinical usability,
data security, and scalability. These priorities ensure
the system functions effectively in a fast-paced clini-
cal environment while adhering to strict data protec-
tion standards. CSGDB features a modular architec-
ture that allows for future expansions, enabling the
implementation of additional functionalities or sys-
tem integrations without disrupting core operations.
The primary design objectives for CSGDB are
to facilitate comprehensive data entry for patient
records, support straightforward data retrieval and
analysis, and ensure compliance with legal require-
ments for patient data security, such as GDPR stan-
dards. The system utilizes a modern technology stack
that includes ElectronJS for cross-platform applica-
tion development, ReactJS for creating a responsive
and intuitive user interface, SQLite as the primary
database for reliable data storage and portability, and
NodeJS as the backend framework. This combina-
tion ensures that CSGDB is lightweight and high-
performing, providing clinicians with an efficient and
fast system. It is easily deployable on various oper-
ating systems used in healthcare facilities, further en-
hancing its usability.
The interface design enhances clinical usability by
prioritizing ease of navigation and efficient data en-
try, allowing clinicians to input and access patient in-
formation quickly. The layout minimizes cognitive
load, featuring customizable fields tailored to salivary
gland tumors. These fields capture essential clinical
and demographic details in an organized manner. This
level of customization promotes consistency in data
collection, which is beneficial for tracking patients
over time and analyzing outcomes.
Data security is paramount in CSGDB, given the
sensitive nature of patient records. The system in-
corporates encryption mechanisms, access control,
and anonymization protocols to protect patient iden-
tities and ensure that only authorized personnel can
access or modify data. This approach safeguards
privacy and supports data sharing for research pur-
poses, where anonymized patient data can drive in-
sights while maintaining confidentiality.
Lastly, scalability is a key design consideration,
allowing CSGDB to grow alongside advancements in
clinical informatics and data management. The sys-
tem’s architecture and technology choices support ef-
ficiently handling an increasing volume of data. Fu-
ture enhancements, such as machine learning mod-
els for predictive analysis, can be implemented with
minimal changes to the existing infrastructure. This
flexibility ensures that CSGDB remains a valuable
resource for both clinical practice and research over
time.
3.2 Architecture Overview
CSGDB is designed as a client-server (Oluwatosin,
2014) application (see Figure 1), providing a clear
separation between data management on the server
side and user interaction on the client side. The server
Czech Salivary Gland Database
707

manages data storage and processing, while the client
provides healthcare professionals with intuitive ac-
cess to the system’s functionalities through a user-
friendly interface.
As illustrated in Figure 1, the CSGDB system ar-
chitecture is built around a relational database back-
end that handles both structured medical data (e.g.,
patient demographics, tumor classifications) and un-
structured notes (e.g., clinician observations). This
database forms the core of the application, ensuring
reliable and secure data storage. An API layer facili-
tates communication between the database and client
applications, providing seamless access to data.
We designed the client side to provide a re-
sponsive, cross-platform user experience that func-
tions smoothly across different operating systems.
The client-side application is structured around sev-
eral key components to address the diverse needs
of healthcare professionals. These include the pa-
tients list component, which displays a searchable and
sortable list of patients; the studies list component,
which organizes studies and their associated patient
data for efficient navigation; the add patient com-
ponent, allowing users to add new patient records
seamlessly; the add study component, facilitating the
creation of new studies with relevant patient associ-
ations; and the Kaplan-Meier component, enabling
clinicians to generate survival curves for advanced
data analysis.
On the server side, the back-end processes incom-
ing requests, manages interactions with the database,
and handles the application’s core business logic. One
of the most essential modules in the back-end is the
encryption module, which is responsible for encrypt-
ing all the sensitive patient data.
Communication between the front-end and back-
end is achieved with the Inter-Process Communica-
tion module. Its purpose is to process user requests
from the front-end, then call appropriate functions on
the back-end and provide a response with the result of
the desired operation.
In addition to the core client-server architecture,
the system’s modular structure supports robust scala-
bility. Future enhancements, such as adding predic-
tive analytics or extending data processing capabili-
ties, can be seamlessly integrated. The clear separa-
tion of responsibilities within the architecture ensures
the secure handling of sensitive patient data.
3.3 Data Security and Anonymization
Data security within CSGDB is implemented through
robust encryption, access control, and anonymization
protocols. All sensitive patient data are encrypted
upon entry and stored securely in the database, en-
suring protection against unauthorized access. To
further safeguard patient privacy, pseudonymization
techniques are applied so that direct identifiers are
separated from clinical information, minimizing the
risk of re-identification. Access control is managed
through two-factor authentication, allowing only au-
thorized users to view or edit specific data. These
measures collectively ensure a high data security and
privacy standard, enabling secure handling of sensi-
tive medical information in clinical and research set-
tings.
4 KEY FUNCTIONALITIES OF
CSGDB
CSGDB includes several key functionalities designed
to streamline the data management process for clini-
cians and researchers, enabling better data utilization
and visualization in clinical decision-making.
Figure 2: Screenshot of Czech Salivary Gland Database
User Interface (Brierley et al., 2017).
4.1 User Interface and Experience
Our main goal for the CSGDB user interface was to
make it straightforward for clinical staff to access. We
devised a simple design that included two main com-
ponents to accomplish this. The main menu is on the
program window’s left side, allowing users to access
the application’s core capabilities immediately. The
corresponding view for the chosen menu option is dis-
played in the center of the application’s windows (see
Figure 2). This layout enables users to navigate be-
tween each functionality easily.
4.2 Updating Patient Records
Updating patient records is essential for medical pro-
fessionals. Currently, most databases rely on retro-
HEALTHINF 2025 - 18th International Conference on Health Informatics
708

spective data collection, meaning that patient infor-
mation is recorded on a specific date—typically the
date the patient is entered into the system—without
any subsequent updates. Our application addresses
this limitation by enabling the editing of patient
records with a streamlined, user-friendly process.
The update begins when the user opens the pa-
tient’s list component, displaying all patient records.
The user can then locate the specific patient they wish
to edit using search and filtering functionalities. Once
the desired patient is identified, a single click on the
edit button enables editing mode. In this mode, the
user can modify the patient’s details.
The interface provides two options upon complet-
ing the edits. If the user is satisfied with the changes,
they can click the save button, which commits the up-
dated data to the database. Alternatively, if the user
decides to discard the changes, they can click the undo
button, which reverts the record to its previous state.
4.3 Application Security
Because clinical data is one of the most sensitive
pieces of information about anyone’s life, it is nec-
essary to secure it properly. Our application en-
crypts all sensitive information about patients with
AES encryption (Daemen, 1999), (Rijmen and Dae-
men, 2001). To log in to the application, the user
must provide his password simultaneously with the
encryption key; otherwise, access to the application is
not granted.
4.4 Data Filtration
The ability to filter patients based on defined factors is
critical for effectively utilizing clinical datasets. Cur-
rently, the application supports filtration based on pa-
rameters such as the type of impacted salivary gland,
the therapy administered, and the histopathological
classification. The system’s modular design facili-
tates the addition of new filtering criteria based on
user requirements or emerging research needs.
Figure 3: Filtration menu.
The filtration menu, accessible within the patient’s
list component and illustrated in Figure 3, allows
users to configure and apply filters based on vari-
ous criteria. Users can also reset all applied filters
to their default state. These functionalities enhance
the system’s flexibility, supporting targeted data anal-
ysis, which proves especially valuable in clinical and
research contexts.
4.5 Visualization and Analytics
Figure 4: Generated Kaplan-Meier curve.
The application offers users a tool to generate Kaplan-
Meir (Kaplan and Meier, 1958) curves. Currently,
there are two possible curve variations. The first rep-
resents survival probability, while the second depicts
recurrence likelihood.
Patients for the curve dataset are chosen based on
their histopathological type. Multiple Kaplan-Meier
curves can be displayed in the same chart, allowing
quick visual data comparisons between patients with
certain histology classifications. (Goel et al., 2010)
Figure 4 shows the Kaplan-Meier survival curves
for patients across three histopathological groups
listed at the top of the graph. The X-axis represents
time in years, and the Y-axis indicates the probability
of survival.
4.6 Data Back-Up and Import/Export
The CSGDB offers robust tools for managing data
integrity and interoperability. Users can perform
database back-ups, a critical feature given the ex-
tended timeframes often required for comprehensive
data collection. These back-ups are securely stored
and encrypted, ensuring that sensitive data remains
protected even in unexpected damage to the user’s
machine. Each back-up file can be restored within
the application by providing the correct password and
encryption key, ensuring only authorized users can
access the data. While the application does not cur-
Czech Salivary Gland Database
709
rently include tools for advanced statistical analysis, it
enables data exchange through an import/export fea-
ture. Users can export patient and study data into Ex-
cel files, allowing for detailed statistical processing
in external software. This functionality ensures clin-
icians and researchers can leverage specialized ana-
lytical tools without compromising data security or
usability.
Understanding the sensitive nature of patient
data, the application provides an export option that
anonymizes all personal patient data. This feature is
particularly useful when medical professionals share
their research data, ensuring patient privacy is always
protected.
It is also possible to import the Excel file with data
into CSGDB, which enables sharing of the collected
data between application instances.
5 IMPLEMENTATION AND
DEPLOYMENT
study
PK
id int NOT NULL
study_name TEXT NOT NULL
study_type INT NOT NULL
is_in_study
PK
id INT NOT NULL
FK1
id_study INT NOT NULL
FK2
id_patient INT NOT NULL
patient_type INT NOT NULL
password
PK
id INT NOT NULL
password TEXT
encryption_enabled TINYINT
submandibular_patient
PK
id INT NOT NULL
form_type INT NOT NULL
name TEXT
surname TEXT
// other basic information
...
// gland specific information
...
parotid_patient
PK
id INT NOT NULL
form_type INT NOT NULL
name TEXT
surname TEXT
// other basic information
...
// gland specific information
...
sublingual_patient
PK
id INT NOT NULL
form_type INT NOT NULL
name TEXT
surname TEXT
// other basic information
...
// gland specific information
...
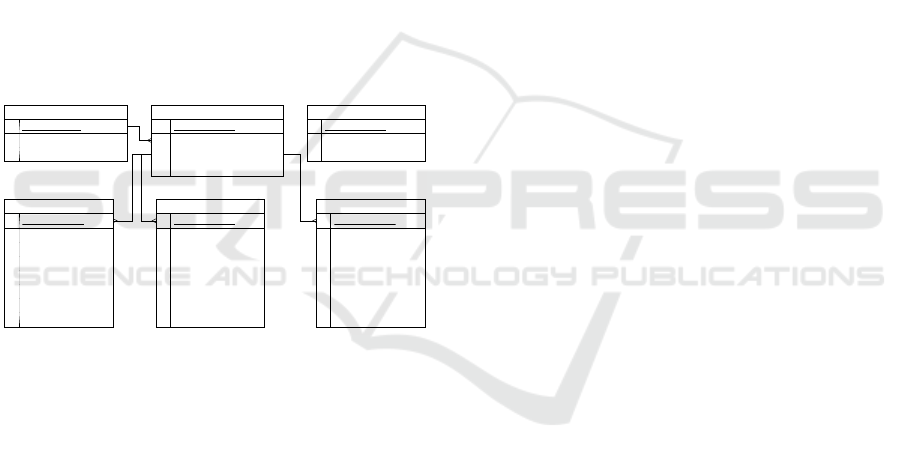
Figure 5: ERA Model of CSGDB.
The CSGDB prioritizes reliability and scalability,
leveraging a modern tech stack to ensure secure, ef-
ficient, and compliant operations within a clinical en-
vironment.
5.1 Technology Stack and Data Model
The CSGDB is built on a modern technology stack
that provides secure, scalable, and efficient data man-
agement. It employs an SQLite relational database
for data storage, ensuring reliability and flexibility in
handling clinical data. The backend is implemented
using a Node.js-based API, while the front-end inter-
face is developed with ReactJS, providing a respon-
sive and user-friendly experience.
The underlying database schema is structured
to meet the specific needs of salivary gland tumor
data management. Figure 5 illustrates the Entity-
Relationship-Attribute (ERA) model of the database,
which comprises six tables, one for each gland that
may be affected by a tumor. Each study can include n
patients, and each patient can participate in n studies,
reflecting the complex relationships inherent in clini-
cal datasets.
Additionally, the database includes a password
table to store hashed passwords and encryption-
related metadata. Users can enable or disable encryp-
tion during the application’s initial setup. This feature
supports testing with fictitious data while maintaining
security for actual clinical data.
5.2 Deployment Context
CSGDB was deployed at the University Hospital in
Motol as a pilot program. This setting allowed for
real-world testing and validation of the system’s us-
ability and performance.
6 CONCLUSIONS
The Czech Salivary Gland Database (CSGDB) is a
significant contribution to clinical data management
and oncological research. By addressing challenges
such as secure storage, real-time updates, and data
visualization, the platform provides a structured ap-
proach to managing data related to salivary gland tu-
mors. The tailored design of CSGDB facilitates clin-
ical decision-making and research, bridging a critical
gap in the availability of domain-specific information
systems.
The development of CSGDB involved integrating
advanced technological components, including AES
encryption for data security, modular architecture for
extensibility, and tools for visualizing survival out-
comes. Its deployment in a clinical setting demon-
strates the system’s applicability and reliability un-
der real-world conditions. These achievements un-
derscore the value of combining technological inno-
vation with domain-specific requirements in develop-
ing medical information systems.
Future work will focus on extending the database
to encompass additional rare tumor types, thereby in-
creasing its applicability to a broader range of onco-
logical studies. The planned integration of predictive
analytics and large language models will further en-
hance the system’s capacity to provide insights, con-
tributing to advancements in personalized medicine.
These directions align with the broader objectives
of improving patient outcomes and facilitating high-
quality research in head and neck oncology.
This work exemplifies the role of targeted
databases in advancing clinical research and practice,
HEALTHINF 2025 - 18th International Conference on Health Informatics
710

offering a model for future systems to address specific
challenges in medical data management and analysis.
ACKNOWLEDGEMENTS
This work was supported by Grant No. SGS-2022-
016 Advanced methods of data processing and analy-
sis.
REFERENCES
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L.,
Soerjomataram, I., and Jemal, A. (2024). Global
cancer statistics 2022: Globocan estimates of inci-
dence and mortality worldwide for 36 cancers in 185
countries. CA: A Cancer Journal for Clinicians,
74(3):229–263.
Brierley, J. D., Gospodarowicz, M. K., and Wittekind, C.
(2017). TNM Classification of Malignant Tumours.
John Wiley & Sons, Ltd, UICC, eighth edition.
Daemen, J. (1999). Aes proposal: Rijndael.
Ferlay, J., Ervik, M., Lam, F., Laversanne, M.,
Colombet, M., Mery, L., Pi
˜
neros, M., Znaor,
A., Soerjomataram, I., and Bray, F. (2024).
Global cancer observatory: Cancer today.
Available from: https://gco.iarc.who.int/today,
https://gco.iarc.who.int/media/globocan/factsheets/
populations/208-denmark-fact-sheet.pdf,
https://gco.iarc.who.int/media/globocan/factsheets/
populations/203-czechia-fact-sheet.pdf.
Friedman, S. and Negoita, S. (2024). History of the surveil-
lance, epidemiology, and end results (seer) program.
JNCI Monographs, 2024(65):105–109.
Goel, M. K., Khanna, P., and Kishore, J. (2010). Under-
standing survival analysis: Kaplan-meier estimate. In-
ternational journal of Ayurveda research, 1(4):274.
Hankey, B. F., Ries, L. A., and Edwards, B. E. (1999). The
surveillance, epidemiology, and end results program:
A national resource. Cancer Epidemiology, Biomark-
ers & Prevention, 8:1117–1121.
Kalfe
ˇ
rt, D., Jel
´
ınek, V., Nov
´
y, P., and Br
˚
uha, P. (2024).
Czech salivary gland database - a computer software
for managing a database of patients with malignant
tumors of the major salivary glands. In 10th Czech-
Slovak Congress, the 85th Congress of the Czech ENT
Society of JEP, and the 70th Congress of the Slovak
Society for ENT and Head and Neck Surgery.
Kaplan, E. L. and Meier, P. (1958). Nonparametric esti-
mation from incomplete observations. Journal of the
American Statistical Association, 53(282):457–481.
Oluwatosin, H. S. (2014). Client-server model. IOSR Jour-
nal of Computer Engineering, 16(1):67–71.
Overgaard, J., Jovanovic, A., Godballe, C., and Eriksen, J.
(2016). The danish head and neck cancer database.
Clinical Epidemiology, 8:491–496.
Rijmen, V. and Daemen, J. (2001). Advanced encryption
standard. Proceedings of federal information process-
ing standards publications, national institute of stan-
dards and technology, 19:22.
Seethala, R. R., Altemani, A., Ferris, R. L., Fonseca, I.,
Gnepp, D. R., Ha, P., Nagao, T., Skalova, A., Sten-
man, G., and Thompson, L. D. R. (2018). Carcino-
mas of the Major Salivary Glands Histopathology Re-
porting Guide. International Collaboration on Cancer
Reporting, Sydney, Australia, 1st edition.
Thompson, L. D. R., Bishop, J. A., Hyrcza, M. D., Ihrler,
S., Leivo, I., Nagao, T., Rupp, N. J., Skalova, A.,
Stenman, G., Vander Poorten, V., van Herpen, C., and
Helliwell, T. (2024). Carcinomas of the Major Sali-
vary Glands Histopathology Reporting Guide. Inter-
national Collaboration on Cancer Reporting, Sydney,
Australia, 2nd edition.
Czech Salivary Gland Database
711
