
From Polar Bears to People: The Role of Ethnic Genetic Variation in
Thermoregulation and Heat-Related Health Risk
Alexandra Baumann
1,∗ a
, Jakob Thiel
2,∗ b
, Nina Haffer
3,4 c
, Shailendra Gupta
1 d
and
Markus Wolfien
2,5 e
1
Department of Systems Biology and Bioinformatics, University of Rostock, Rostock, Germany
2
Institute for Medical Informatics and Biometry, Faculty of Medicine and University Hospital Carl Gustav Carus, TUD
Dresden University of Technology, Dresden, Germany
3
Berlin Institute of Health at Charit
´
e - Universitatsmedizin Berlin, Germany
4
Charit
´
e - University Medicine Berlin, Germany
5
Center for Scalable Data Analytics and Artificial Intelligence (ScaDS.AI), Dresden/Leipzig, Dresden, Germany
fi
Keywords:
Heat Illness, Heat Susceptibility, Climate Change, Ethnicity, Genetic Association.
Abstract:
As climate change increases the frequency and severity of acute heat events, it is crucial to determine fac-
tors for appropriate healthcare strategies and predictive models. Previously, it was stated that socioeconomic
factors primarily play a role in heat-related illness risk. Analogous to the polar bear’s unique adaptations
to the cold, humans exhibit distinct genetic traits shaped by their migration to diverse climates. This posi-
tion paper hypothesizes that genetic differences among human ethnic groups, in addition to socioeconomic
and other factors, also contribute to variations in thermoregulation and influence susceptibility to heat-related
diseases. To understand genetic adaptations across human ethnicities (initially European and African), we
propose a genetic association analysis of single nucleotide polymorphisms (SNPs) in genes associated with
thermoregulation. An assessment of changes in thermoregulation gene regulation networks will be possible
by conducting a functional pathway analysis. Expected outcomes include identifying differences in SNP dis-
tributions of thermoregulation-associated genes across ethnicities. Challenges such as the underrepresentation
of African populations in genomic databases must also be addressed. This research aims to provide a foun-
dational understanding of genetic contributions to heat adaptation, guiding the development of personalized,
equitable healthcare responses to climate-induced heat stress.
1 BACKGROUND
1.1 The Polar Bear’s Struggle with
Climate Change
Climate change is driving widespread environmental
shifts, notably the melting of the ice caps and an over-
all increase of global temperatures. The polar bear,
native to arctic regions, faces challenges as its habi-
tat warms beyond its evolutionary adaptations. Un-
a
https://orcid.org/0009-0003-8946-8741
b
https://orcid.org/0009-0007-1951-1678
c
https://orcid.org/0000-0002-9541-2811
d
https://orcid.org/0000-0002-3470-3260
e
https://orcid.org/0000-0002-1887-4772
∗
Shared first authorship.
like other bear species, the polar bear differentiates
by diverse characteristics, such as adaptation to colder
temperatures by dual-layered fur or slip-proof feet
(Welch et al., 2014). Imagine a polar bear visiting
black bears or grizzly bears in warmer regions, where
it lacks adaptations for heat. While black bear’s ears
are large for heat dissipation, the ears of the polar bear
are smaller and the they have a thick layer of subcuta-
neous fat (Rinker et al., 2019). The polar bear is also
bigger than the other bear species in warmer regions.
This can be explained by a rule proposed by the sci-
entist Carl Bergmann, who discovered that these size
variations relate to the surface area-to-body mass ra-
tio (Bergmann, 1848). Smaller body size is typical in
populations near the equator, while larger body size is
more common in colder regions.
Baumann, A., Thiel, J., Haffer, N., Gupta, S. and Wolfien, M.
From Polar Bears to People: The Role of Ethnic Genetic Variation in Thermoregulation and Heat-Related Health Risk.
DOI: 10.5220/0013256100003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 655-660
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
655

1.2 Human Heat Adaptations Around
the Globe
This natural variation among bears serves as a vivid
analogy: just as bear species adapt to specific cli-
mates, human population may also exhibit diverse
adaptations in resilience to heat - an increasingly im-
portant consideration in our warming world. Cli-
mate adaptations affect not only physical character-
istics but also health factors, such as birth weight
and fetal growth, in populations across diverse cli-
mates (Lambert et al., 2008). As humans migrated
across different climates, it was necessary to slowly
adapt to the environment over multiple generations,
which led to phenotypic variation across ethnicities.
Research on thermoregulation in different ethnici-
ties suggests both genotypic and phenotypic adapta-
tions to heat (Taylor, 2006; Lim, 2020). Since 2006,
genetic variation has been proposed as an adaptive
mechanism for heat tolerance, with regulatory pro-
cesses controlling individual heat responses (Taylor,
2006). Observable adaptations include skeletal mor-
phology and traits like nose shape, hair type, and
lip structure, with one of the most prominent being
skin and eye color (Lambert et al., 2008). Often,
multiple genetic variants collectively influence a sin-
gle phenotype, highlighting the polygenic nature of
these adaptations (Cort
´
es et al., 2020). The study by
Huang et al. identified 299 single nucleotide poly-
morphisms (SNPs) differing across populations, pri-
marily in genes influencing skin and eye color, sug-
gesting a genetic adaptation to environmental factors
such as increased heat and UV exposure (Huang et al.,
2015). For instance, melanin in the skin provides UV
protection for people living in regions closer to the
equator, while depigmentation enhances vitamin D3
synthesis in populations living farther north (Lambert
et al., 2008). These adaptations also impact metabolic
traits. Metabolic rates vary across populations, and
epigenetic factors may contribute to thermoregula-
tory differences (Cramer et al., 2022). Cold tolerance
appears to be higher among individuals from colder
regions, while those from tropical regions may ex-
hibit a reduced response to heat. Natural selection
likely favored thermoregulatory mutations, such as
those supporting heat production in northern popula-
tions (Lambert et al., 2008). Differences in metabolic
rate, subcutaneous fat levels, thyroid activity, and mi-
tochondrial DNA (mtDNA), may all contribute to in-
creased metabolic heat production in colder climates
(Lambert et al., 2008). Furthermore, molecular differ-
ences can influence protein functions and even drug
responses across populations with implications for
treatments related to heat-induced health conditions
(Duello et al., 2021). Studying these diverse adap-
tations may help inform healthcare practices that ad-
dress climate-related health risks more effectively.
1.3 Impact of Ethnicity and
Socioeconomics on Heat-Related
Illness
Apart from region-specific heat adaptations, climate
change is increasing the frequency and intensity of
acute heat events (WHO, 2023). Heat impacts health
in various ways, causing both direct and indirect ef-
fects. Directly, exposure to high temperatures can
result in heat-related illnesses, such as heat stroke,
dehydration, or heat collapse (Xu et al., 2023). In-
directly, heat can increase the risk of severe events
like heart attacks and is associated with higher mor-
tality rates among vulnerable populations, including
cancer patients (H
¨
using et al., 2024). The rise in
acute heat events also places a considerable strain on
healthcare systems (WHO, 2023). One response to
this challenge is the development of predictive mod-
els to forecast hospital resource needs, combining cli-
mate and medical data (Thiel et al., 2024). Adding
patient-specific characteristics as risk factors, such as
age and gender, can improve the accuracy of these
models (Cheng et al., 2019).) Studies by Berberian
et al. (2022) and Jackson et al. (2022) indicate that
people with darker skin may be at higher risk of heat-
related illnesses (Berberian et al., 2022; Jackson et al.,
2022). Both studies highlight that socioeconomic fac-
tors often underlie this increased risk. However, ge-
netic factors influencing thermoregulation may also
contribute to these differences, as discussed above.
This suggests that ethnic background could be an es-
sential factor in developing predictive models for heat
illnesses, providing a more comprehensive approach
in protecting diverse populations.
This position paper presents the hypothesis that it
would be valuable to investigate whether genetic dif-
ferences between individuals of different ethnicities
can be associated with thermoregulation and, conse-
quently, the risk of heat-related illnesses. By investi-
gating these genetic factors, the study aims to identify
potential contributors to heat-related illness suscepti-
bility, ultimately supporting the development of pre-
cise prevention measures and predictive models.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
656

2 PROPOSED APPROACH ON
ANALYZING THE IMPACT OF
ETHNICITY ON
THERMOREGULATION
The impact of ethnic genetic variation on heat-related
diseases has to be inspected to test the hypothesis. Ex-
isting literature on thermoregulation-associated genes
and gene regulation networks provides a foundation
for this investigation, offering insights into pathways
and mechanisms in heat resilience and adaptation.
Heat regulation plays a crucial role in cellular house-
keeping and homeostasis as well as in stress response
(Charlebois et al., 2018). Further processes and bi-
ological components that might be of interest are
thermo-receptors, thermo-sensitive neurons, or lipol-
ysis (Valero et al., 2014). Diverse databases offer
a systematic framework to identify genes related to
biological processes such as Gene Ontology (GO)
(Aleksander et al., 2023; Ashburner et al., 2000),
or even associations between diseases and variants
like Disgenet (Pinero et al., 2017). Relevant GO
terms, amongst others, include ”heat acclimation”
(GO:0010286), ”response to heat” (GO:0009408),
”cellular response to heat” (GO:0034605), ”heat
generation” (GO:0009409), ”temperature homeosta-
sis” (GO:0001659), ”circadian temperature home-
ostasis” (GO:0003052), or ”sweat gland develop-
ment” (GO:0061114). For example, the GO term
”heat acclimation” is linked to 366 genes and gene
products across all organisms in the database, of
which six are human genes. Similarly, ”temperature
homeostasis” is associated with 264 genes across all
organisms, including 34 human genes. As a start-
ing point, a genetic association analysis could be con-
ducted utilizing common SNP databases such as db-
SNP (Sherry et al., 1999) or gnomAD (Karczewski
et al., 2020) to identify SNPs in thermoregulation-
related genes, with allele frequencies analyzed ac-
cording to ethnicity. Initially, the primary comparison
groups will be Europeans (non-Finnish) and Africans
(/African Americans), as these ethnicities are already
represented in common SNP databases, enabling a
robust baseline analysis of allele frequency differ-
ences. To predict the potential effect of each vari-
ant on gene function, including regulatory roles, the
Ensembl Variant Effect Predictor (VEP) tool will be
applied (McLaren et al., 2016). The main focus will
be on SNPs in protein-coding regions. A preliminary
analysis focusing on 40 genes associated with the GO
terms ”heat acclimation” and ”temperature homeosta-
sis” revealed 288 variants from the gnomAD v4.1.0
database with allele frequency differences greater
than 0.5 % between the two before-mentioned pop-
ulations. First filtering steps excluded variants in in-
tronic (not splice-relevant) and UTR regions or vari-
ants with an allele frequency below 0.5 % in the pop-
ulation with the maximum allele frequency. Ten vari-
ants with highest difference between the two popu-
lation allele frequencies in Europeans (non-Finnish)
and Africans (/African Americans) are depicted in
(Table 1).
There are some noticeable differences between
variant allele frequencies across the two populations
in genes associated to thermoregulation. Both popu-
lations are represented with either higher or lower al-
lele frequencies for the variants. Synonymous as well
as missense variants or splice-associated ones can be
found in this first view. The variants have to inspected
for their relevance in gene functions. In this first
consideration, only exonic and splice region variants
were investigated. However, as SNPs in intronic and
intergenic regions can influence transcription regula-
tion or affect regulatory RNAs (such as micro RNAs),
those SNPs will be examined as well. This analysis
will help to simulate and assess changes in gene reg-
ulation networks involved in thermoregulation. Ad-
ditionally, functional pathway analysis tools, such as
KEGG (Kanehisa and Goto, 2000) or Reactome (Fab-
regat et al., 2017), could be applied to map these SNPs
within thermoregulation pathways, deepening our un-
derstanding of how variations in these pathways may
contribute to heat resilience or susceptibility. While
the initial focus will be on European and African eth-
nicity, future analyses could be expanded to include
other ethnic groups (such as East Asian or Indigenous
populations) to achieve a more comprehensive view
on thermoregulatory adaptations across diverse envi-
ronments. Incorporating environmental data, such as
historic climate conditions associated with each pop-
ulation, could further contextualize observed genetic
variations as adaptive responses to different climates.
3 EXPECTED INFLUENCES AND
CHALLENGES
We expect to find differences in SNP distributions
between the two ethnicities under study, specifically
within genes associated with thermoregulation. These
SNP variations may contribute to observed differ-
ences in thermoregulation among diverse ethnicities,
particularly between Europeans and Africans in this
analysis. It is also anticipated that some SNPs will
appear in intronic or non-coding regions, which may
not directly impact gene function and could be con-
sidered incidental ”byproducts.” Nevertheless, it re-
From Polar Bears to People: The Role of Ethnic Genetic Variation in Thermoregulation and Heat-Related Health Risk
657
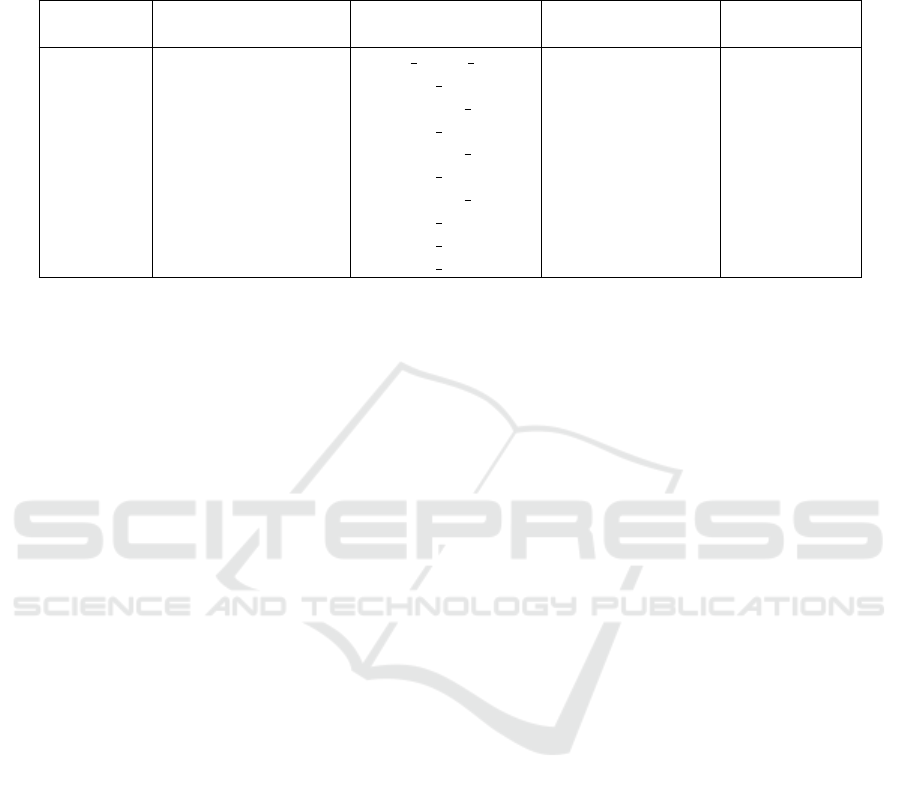
Table 1: Top ten variants in thermoregulation-related genes with highest allele frequency (AF) differences between
African/African American and European (non-Finnish) populations. HGVS nomenclature of variant consequence on cDNA
(c.) or protein (p.) level. Variant Effect Predictor (VEP) annotation of variant region and consequence. Contents extracted
and adapted from gnomAD v4.1.0 (Karczewski et al., 2020).
Gene HGVS Consequence VEP Annotation
AF African/
African American
AF European
(non-Finnish)
STAT3 c.1601-8dup splice region variant 0.6847 0.1937
NAPEPLD p.Asp389Asn missense variant 0.5586 0.9989
DRD2 p.Pro319Pro synonymous variant 0.1168 0.5468
MC3R p.Val44Ile missense variant 0.4284 0.0839
DRD2 p.His313His synonymous variant 0.3622 0.7057
HSPA1A p.Glu110Asp missense variant 0.4176 0.1059
TRPM2 p.Asp1360Asp synonymous variant 0.2808 0.0004
TRPV1 p.Thr469Ile missense variant 0.0741 0.3517
ADRB2 p.Glu27Gln missense variant 0.8239 0.5570
RBBP7 p.Arg37His missense variant 0.1899 0.4445
mains essential to investigate the intronic and non-
coding SNPs, too. They can be located in regula-
tory regions with an effect on transcription rate and
transcript stability or can lead to alternative splicing
(Vaz-Drago et al., 2017). Furthermore, micro RNAs
with key functions in pathway regulations are often
located in intronic regions (Vaz-Drago et al., 2017).
A key challenge, therefore, will be filtering out those
SNPs with a meaningful impact on thermoregulation.
A limitation of this proposed approach is the over-
representation of European populations in genomic
databases, while African populations remain com-
paratively under-studied. This imbalance may affect
the generalizability of the findings. Additionally, ge-
nomic ancestry alone does not fully capture socioe-
conomic diversity within or between African and Eu-
ropean populations. Socioeconomic factors can in-
fluence health outcomes through epigenetic modifica-
tions, which may impact thermoregulation and other
heat-related traits. Further challenges include ac-
counting for factors such as sex, age, and pre-existing
conditions, which are likely to influence thermoregu-
latory responses and the risk of heat-related illnesses.
Analyzing male and female participants separately
may reveal sex-specific effects on thermoregulation,
and considering age groups and health status will be
essential for an in-depth interpretation of the results.
Lastly, the applicability of these findings to clinical
settings may require long-term studies, as epigen-
tic and genetic influences on thermoregulation likely
evolve over time.
4 IMPLICATIONS AND
CONCLUSION
This study underscores the importance of examin-
ing genetic diversity in understanding thermoregula-
tion and susceptibility to heat-related illnesses in ad-
dition to socioeconomic factors. By identifying ge-
netic variations associated with heat resilience and
mapping them across diverse ethnicites, we can de-
velop more precise predictive models and preventive
measures tailored to individual needs. The proposed
approach will support health equity, offering insights
that help to mitigate the disproportionate impact of
climate change.
Just as polar bears are uniquely adapted to cold en-
vironments but face challenges as temperatures rise,
different human ethnicities may too require unique
adaptations as they confront a warming world. Un-
derstanding these genetic differences equips us to de-
velop strategies to support all individuals in the face
of changing climates, preserving health across the di-
versity of humanity.
ACKNOWLEDGEMENTS
We are particularly grateful for the financial support
provided by the Federal Ministry of Education and
Research (BMBF: OLCIR - 02NUK082C, MiHUBx
- 01ZZ2101A).
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
658

REFERENCES
Aleksander, S. A., Balhoff, J., Carbon, S., Cherry, J. M.,
Drabkin, H. J., Ebert, D., Feuermann, M., Gaudet, P.,
Harris, N. L., Hill, D. P., Lee, R., Mi, H., Moxon, S.,
Mungall, C. J., Muruganugan, A., Mushayahama, T.,
Sternberg, P. W., Thomas, P. D., Van Auken, K., Ram-
sey, J., Siegele, D. A., Chisholm, R. L., Fey, P., As-
promonte, M. C., Nugnes, M. V., Quaglia, F., Tosatto,
S., Giglio, M., Nadendla, S., Antonazzo, G., Attrill,
H., dos Santos, G., Marygold, S., Strelets, V., Tabone,
C. J., Thurmond, J., Zhou, P., Ahmed, S. H., Asanit-
thong, P., Luna Buitrago, D., Erdol, M. N., Gage,
M. C., Ali Kadhum, M., Li, K. Y. C., Long, M.,
Michalak, A., Pesala, A., Pritazahra, A., Saverimuttu,
S. C. C., Su, R., Thurlow, K. E., Lovering, R. C., Lo-
gie, C., Oliferenko, S., Blake, J., Christie, K., Cor-
bani, L., Dolan, M. E., Drabkin, H. J., Hill, D. P.,
Ni, L., Sitnikov, D., Smith, C., Cuzick, A., Seager, J.,
Cooper, L., Elser, J., Jaiswal, P., Gupta, P., Jaiswal,
P., Naithani, S., Lera-Ramirez, M., Rutherford, K.,
Wood, V., De Pons, J. L., Dwinell, M. R., Hay-
man, G. T., Kaldunski, M. L., Kwitek, A. E., Lauled-
erkind, S. J. F., Tutaj, M. A., Vedi, M., Wang, S.-J.,
D’Eustachio, P., Aimo, L., Axelsen, K., Bridge, A.,
Hyka-Nouspikel, N., Morgat, A., Aleksander, S. A.,
Cherry, J. M., Engel, S. R., Karra, K., Miyasato,
S. R., Nash, R. S., Skrzypek, M. S., Weng, S., Wong,
E. D., Bakker, E., Berardini, T. Z., Reiser, L., Auch-
incloss, A., Axelsen, K., Argoud-Puy, G., Blatter, M.-
C., Boutet, E., Breuza, L., Bridge, A., Casals-Casas,
C., Coudert, E., Estreicher, A., Livia Famiglietti, M.,
Feuermann, M., Gos, A., Gruaz-Gumowski, N., Hulo,
C., Hyka-Nouspikel, N., Jungo, F., Le Mercier, P.,
Lieberherr, D., Masson, P., Morgat, A., Pedruzzi, I.,
Pourcel, L., Poux, S., Rivoire, C., Sundaram, S., Bate-
man, A., Bowler-Barnett, E., Bye-A-Jee, H., Denny,
P., Ignatchenko, A., Ishtiaq, R., Lock, A., Lussi, Y.,
Magrane, M., Martin, M. J., Orchard, S., Raposo, P.,
Speretta, E., Tyagi, N., Warner, K., Zaru, R., Diehl,
A. D., Lee, R., Chan, J., Diamantakis, S., Raciti, D.,
Zarowiecki, M., Fisher, M., James-Zorn, C., Ponfer-
rada, V., Zorn, A., Ramachandran, S., Ruzicka, L., and
Westerfield, M. (2023). The Gene Ontology knowl-
edgebase in 2023. Genetics, 224(1):iyad031.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D.,
Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K.,
Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P.,
Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese,
J. C., Richardson, J. E., Ringwald, M., Rubin, G. M.,
and Sherlock, G. (2000). Gene Ontology: tool for the
unification of biology. Nature genetics, 25(1):25–29.
Berberian, A. G., Gonzalez, D. J. X., and Cushing, L. J.
(2022). Racial Disparities in Climate Change-Related
Health Effects in the United States. Current Environ-
mental Health Reports, 9(3):451–464.
Bergmann, C. (1848).
¨
Uber die Verh
¨
altnisse der
W
¨
arme
¨
okonomie der Thiere zu ihrer Gr
¨
osse. 3.
Charlebois, D. A., Hauser, K., Marshall, S., and Bal
´
azsi, G.
(2018). Multiscale effects of heating and cooling on
genes and gene networks. Proceedings of the National
Academy of Sciences, 115(45).
Cheng, J., Xu, Z., Bambrick, H., Prescott, V., Wang, N.,
Zhang, Y., Su, H., Tong, S., and Hu, W. (2019). Car-
diorespiratory effects of heatwaves: A systematic re-
view and meta-analysis of global epidemiological ev-
idence. Environmental Research, 177:108610.
Cort
´
es, A. J., L
´
opez-Hern
´
andez, F., and Osorio-Rodriguez,
D. (2020). Predicting Thermal Adaptation by Looking
Into Populations’ Genomic Past. Frontiers in Genet-
ics, 11.
Cramer, M. N., Gagnon, D., Laitano, O., and Crandall,
C. G. (2022). Human temperature regulation under
heat stress in health, disease, and injury. Physiologi-
cal Reviews, 102(4):1907–1989.
Duello, T. M., Rivedal, S., Wickland, C., and Weller, A.
(2021). Race and genetics versus ‘race’ in genetics: A
systematic review of the use of African ancestry in ge-
netic studies. Evolution, Medicine, and Public Health,
9(1):232.
Fabregat, A., Sidiropoulos, K., Viteri, G., Forner, O.,
Marin-Garcia, P., Arnau, V., D’Eustachio, P., Stein,
L., and Hermjakob, H. (2017). Reactome pathway
analysis: a high-performance in-memory approach.
BMC Bioinformatics, 18:142.
Huang, T., Shu, Y., and Cai, Y.-D. (2015). Genetic
differences among ethnic groups. BMC Genomics,
16(1):1093.
H
¨
using, J., Beiderbeck, M., Berressem, S., Kaj
¨
uter, H.,
Stang, A., and Zander, T. (2024). Hitzebedingte
¨
Ubersterblichkeit unter in Nordrhein-Westfalen Kreb-
serkrankten im fortgeschrittenen Stadium. page Do-
cAbstr. 596. German Medical Science GMS Publish-
ing House.
Jackson, P., Larkin, D., Kinnie, K. R., and Aroke, E. N.
(2022). Heat Islands and Chronic Disease: Could
African Americans Be More Vulnerable to Heat-
Related Health Impacts? Journal of National Black
Nurses’ Association : JNBNA, 33(1):33.
Kanehisa, M. and Goto, S. (2000). KEGG: Kyoto Ency-
clopedia of Genes and Genomes. Nucleic Acids Re-
search, 28(1):27.
Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings,
B. B., Alf
¨
oldi, J., Wang, Q., Collins, R. L., Laricchia,
K. M., Ganna, A., Birnbaum, D. P., Gauthier, L. D.,
Brand, H., Solomonson, M., Watts, N. A., Rhodes,
D., Singer-Berk, M., England, E. M., Seaby, E. G.,
Kosmicki, J. A., Walters, R. K., Tashman, K., Far-
joun, Y., Banks, E., Poterba, T., Wang, A., Seed, C.,
Whiffin, N., Chong, J. X., Samocha, K. E., Pierce-
Hoffman, E., Zappala, Z., O’Donnell-Luria, A. H.,
Minikel, E. V., Weisburd, B., Lek, M., Ware, J. S.,
Vittal, C., Armean, I. M., Bergelson, L., Cibulskis, K.,
Connolly, K. M., Covarrubias, M., Donnelly, S., Fer-
riera, S., Gabriel, S., Gentry, J., Gupta, N., Jeandet,
T., Kaplan, D., Llanwarne, C., Munshi, R., Novod, S.,
Petrillo, N., Roazen, D., Ruano-Rubio, V., Saltzman,
A., Schleicher, M., Soto, J., Tibbetts, K., Tolonen, C.,
Wade, G., Talkowski, M. E., Neale, B. M., Daly, M. J.,
and MacArthur, D. G. (2020). The mutational con-
straint spectrum quantified from variation in 141,456
humans. Nature, 581(7809):434–443.
From Polar Bears to People: The Role of Ethnic Genetic Variation in Thermoregulation and Heat-Related Health Risk
659

Lambert, M. I., Mann, T., and Dugas, J. P. (2008). Ethnic-
ity and Temperature Regulation. Medicine and sport
science, 53.
Lim, C. L. (2020). Fundamental Concepts of Human Ther-
moregulation and Adaptation to Heat: A Review in
the Context of Global Warming. International Jour-
nal of Environmental Research and Public Health,
17(21):7795.
McLaren, W., Gil, L., Hunt, S. E., Riat, H. S., Ritchie,
G. R. S., Thormann, A., Flicek, P., and Cunning-
ham, F. (2016). The Ensembl Variant Effect Predictor.
Genome Biology, 17(1):122.
Pinero, J., Bravo, A., Queralt-Rosinach, N., Gutierrez-
Sacrist
´
an, A., Deu-Pons, J., Centeno, E., Garcia-
Garcia, J., Sanz, F., and Furlong, L. I. (2017). Dis-
GeNET: a comprehensive platform integrating in-
formation on human disease-associated genes and
variants. Nucleic Acids Research, 45(Database
issue):D833–D839.
Rinker, D. C., Specian, N. K., Zhao, S., and Gibbons,
J. G. (2019). Polar bear evolution is marked by rapid
changes in gene copy number in response to dietary
shift. Proceedings of the National Academy of Sci-
ences of the United States of America, 116(27):13446.
Sherry, S. T., Ward, M., and Sirotkin, K. (1999).
dbSNP—Database for Single Nucleotide Polymor-
phisms and Other Classes of Minor Genetic Variation.
Genome Research, 9(8):677–679.
Taylor, N. A. S. (2006). Ethnic differences in thermoregu-
lation: Genotypic versus phenotypic heat adaptation.
Journal of Thermal Biology, 31(1):90–104.
Thiel, J., Seim, A., Grummt, S., Nesterow, I., Penesch, F.,
Sedlmayr, M., and Weidner, J. (2024). Tools for op-
timizing healthcare resource allocation in response to
climate impacts and heat action planning. Journal of
Public Health.
Valero, K. C. W., Pathak, R., Prajapati, I., Bankston, S.,
Thompson, A., Usher, J., and Isokpehi, R. D. (2014).
A candidate multimodal functional genetic network
for thermal adaptation. PeerJ, 2:e578.
Vaz-Drago, R., Cust
´
odio, N., and Carmo-Fonseca, M.
(2017). Deep intronic mutations and human disease.
Human Genetics, 136(9):1093–1111.
Welch, A. J., Bedoya-Reina, O. C., Carretero-Paulet, L.,
Miller, W., Rode, K. D., and Lindqvist, C. (2014). Po-
lar Bears Exhibit Genome-Wide Signatures of Bioen-
ergetic Adaptation to Life in the Arctic Environment.
Genome Biology and Evolution, 6(2):433.
WHO (2023). Climate change.
Xu, Z., Watzek, J. T., Phung, D., Oberai, M., Rutherford,
S., and Bach, A. J. E. (2023). Heat, heatwaves, and
ambulance service use: a systematic review and meta-
analysis of epidemiological evidence. International
Journal of Biometeorology, 67(10):1523–1542.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
660
