
Visualizing Medical Coding Practices Using Transformer Models
Tanner Hobson
a
and Jian Huang
b
Department of Electrical Engineering and Computer Science,
University of Tennessee, Knoxville, TN, U.S.A.
Keywords:
Transformer, Medical Codes, Visualization.
Abstract:
In the United States, diagnostic codes are a key component of medical records that document the process of
patient care. It has long been a common belief that there are inherent orders to the sequences of diagnosis
codes in medical records. However, because of the complexities in medical records, there have been few
tools that can automatically distill and make sense of the implicit ordering characteristics of the diagnostic
codes within medical records. With the advent and fast advancement of the Transformer architecture, in this
work we develop and demonstrate a transformer based model named DgPg. DgPg can automatically learn
the patterns in the ordering of diagnostic codes in any given corpus of medical records, for example, those
obtained from the same hospital or those from different hospitals but collected and organized around particular
clinical scenarios. Using DgPg, we can flexibly visualize the coding patterns and context around any particular
diagnostic code. Our results from DgPg further demonstrate that the model learned from one dataset can be
unique to that dataset and, from this respect, confirm that medical coding practices have unique dependencies
on the provider or the clinical scenarios. Our work uses three well known datasets: MIT’s MIMIC-IV dataset,
and CDC’s NHDS and NHCS datasets. Our DgPg transformer models are only 2.5 MB in size. Such compact
footprint enable flexibility in how the models can be deployed for real world use.
1 INTRODUCTION
The Transformer architecture, since first appearing in
2017 (Vaswani, 2017), has had a transformative effect
on language models. A plethora of subsequent works
have led to rapid further development. In 2018, bidi-
rectional encoder representations from transformers
(BERT) (Devlin, 2018) enabled models to understand
context from both the left-to-right and right-to-left di-
rections. In 2020, T5 (Raffel et al., 2020) proposed
that flexible transfer learning can be achieved by treat-
ing all natural language based tasks in a unified text-
to-text format. All of these culminated in the creation
and widespread adoption of LLM models (Brown,
2020), which have fundamentally changed the land-
scape of how AI models are conceived, designed,
used, and perceived.
Our research revolves around medical records,
which are complex, specialized, and messy. In the
United States, a key component in medical records
are the diagnostic codes, which essentially is a “trans-
lation” from unstructured descriptions into structured
labels that characterize the essence of the descrip-
tions. In theory, the sequence of diagnostic codes doc-
a
https://orcid.org/0000-0002-6269-7881
b
https://orcid.org/0000-0002-9288-0505
umented per each patient should provide an accurate,
accountable, and actionable summary that drives the
coordination of that patient’s care.
However, while the step of coding distills infor-
mation, it is a step performed manually, very of-
ten based on experience and intuition. Standardiza-
tion and quality-control are hard. Furthermore, even
though there are often coding manuals to follow, those
manuals have not been designed for ease of use and
hence introduce significant cognitive barriers for the
users, who are already overly task-burdened and at
risk of workplace burnout. Worse yet, different hos-
pital, or different units within the same hospital, may
have differing coding practices due to nuances in their
unique work processes, as well as clinicians’ personal
preferences in how they work.
Due to these reasons of variation, even though
most medical professionals can agree that there must
be inherent patterns among the coding sequences, few
can articulate those patterns with confidence. In addi-
tion, even though coding manuals exist, those cannot
reflect the reality of nuanced patient care and prac-
tical constraints faced by each clinician. From this
respect, no one person truly knows the common cod-
ing practices. In result, it’s hard for a hospital’s lead
physicians and administrators to have a shared under-
Hobson, T. and Huang, J.
Visualizing Medical Coding Practices Using Transformer Models.
DOI: 10.5220/0013257800003905
In Proceedings of the 14th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2025), pages 725-732
ISBN: 978-989-758-730-6; ISSN: 2184-4313
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
725

standing of how patient care is carried out at their hos-
pital. This further hampers quality control, process
improvement, or burnout prevention.
In this work, we hypothesize that the Transformer
architecture can be used to build an AI model to learn
the most representative patterns in coding sequences,
automatically and purely from a corpus of medi-
cal records. Our prototype called DgPg (“Diagnosis
Progression”) is successful. We show that the learned
patterns can be visualized flexibly as AI-generated
visual summaries of the representative coding prac-
tices. The trained transformer models within DgPg
are about 2.5 MB in size. DgPg have been trained on
three publicly accessible datasets: MIMIC-IV (John-
son et al., 2023), NHDS (US CDC NCHS, 2010), and
NHCS (US CDC NCHS, 2020).
Our contributions in this paper are as follows.
To the best of our knowledge, (1) DgPg is the first
to demonstrate that the Transformer architecture can
learn representative patterns in the code sequences
in real-world medical records. (2) This paper is
also the first work that uses AI generated visual-
izations to serve as visual summaries of representa-
tive patterns within a dataset, without the otherwise
lengthy process of requiring a human user to explic-
itly go through the raw data to select features or create
human-provided labels.
We summarize background in Section 2 and
present our research in Section 3. Results from our
experiments and examples of our visualization are in
Section 4. We conclude in Section 5.
2 BACKGROUND
DgPg’s scope is to create organization-level analytics
using an AI model that can learn representative cod-
ing practices within medical records and help hospital
administrators gain operational insights. While that
organizational analytics scope of research is new, we
have drawn inspirations from a plethora of prior re-
search that have analyzed medical records using AI
methods for various clinical use cases.
Some of the best clinical use cases include: to
predict patient readmission and in-hospital mortal-
ity (Hsu et al., 2020), to predict occurrences of sep-
sis hours before human clinicians are able to (Henry
et al., 2015), early detection of diabetic retinopa-
thy (Abr
`
amoff et al., 2018), and cardiovascular dis-
ease diagnosis and prediction (Krittanawong et al.,
2017). Besides clinical outcomes, clinical cost has
been studied too. For example, to correlate a patient’s
medical records across multiple visits in order to re-
veal patterns of patients developing conditions that re-
quire costly procedures (Malhotra et al., 2015).
Specific to sequences pattern mining from medi-
cal records, past work have centered around inter visit
patterns, i.e. across multiple visits by the same pa-
tient. In a large part because there is then a natural
time variable. The most typical data structure used
for modeling is graph (Malhotra et al., 2015). To the
best of our knowledge, our work is the first to examine
representative sequence patterns within the records of
a single visit by a patient.
The medical coder’s job is to understand the im-
plicit reasons behind diagnoses and procedures (Hu
et al., 2021). This job is time consuming. Kim et
al. report that inpatient coders achieve a rate of 2.5
medical charts per hour (Kim and Ganapathi, 2021).
The job is also cognitively intensive, because medi-
cal records involve large sets of synonymous words;
there is rarely a single word that is used to describe
one particular concept (Koleck et al., 2021).
Past researchers have incorporated external
knowledge banks, such as Wikipedia (Bai and
Vucetic, 2019) and UMLS (Chanda et al., 2022)
in an effort to improve AI’s coding capabilities.
In a related manner, not all parts of patient visit
summaries are equally important; using a restricted,
high-quality set of text is more effective than using
all available text (Hsu et al., 2020). Incorporating
attention, a means of assigning importance to dif-
ferent parts of the text, has also become a common
method (Mullenbach et al., 2018).
Lastly, it is worth noting that the direct use case of
DgPg is not to improve medical coding or clinical pre-
dictions. Instead, our use case is to let AI curate how
a complex process is actually implemented in reality
based on data. The ensuing insights can be used for
a variety of purposes. A relevant example of this use
scenario is our prior work, eCamp (Raji et al., 2018),
which is a system that learns and visualizes how uni-
versity students navigate through 100s of potential de-
gree programs at a typical major research university
in the US. All purely based on anonymized electonic
student records. The insights revealed by eCamp can
be used generally by university administrators, aca-
demic advisors, faculty, students, and parents for their
different purposes. We intend DgPg’s insights about
each hospital’s coding practices to have a similar level
of generality and can help disparate stakeholders in
the hospital ecosystem as well. The work of eCamp
predated transformers. To this end, DgPg is the first
attempt of using transformers for such organization-
level analytics.
Next, we describe the design process and technical
details of DgPg in Section 3.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
726

3 THE DgPg TRANSFORMER
MODEL
3.1 Overview
Scope of Use. Every year the United States Center
of Medicare & Medicaid Services (CMS) publishes
the official ICD-10-CM (International Classification
of Disease) coding guidelines (CMS, 2024), which
details specific conventions and rules that guide cod-
ing and sequencing of codes. The sequence of codes
can be due to temporal ordering, etiological ordering,
or situation-specific guidelines.
The codes and sequencing of codes are both sig-
nificant. It’s fraudulent to change coding for the pur-
pose of increasing medical claim payment. Any re-
vision of coding must be done with proper support
of documentation, including documentation of causal
relationship between codes and medical events.
While it is possible, and common practice, for a
human expert to read through the complete medical
records of a patient’s visit to audit each instance of
the coding practice, a computational tool is needed to
systematically understand the coding practice shared
among a unit of healthcare providers. DgPg’s scope
of use is in this context. While DgPg’s overall scope
can include all medical codes, we have chosen to start
with diagnostic (DX) codes in this work to reduce the
complexities.
Design Goals. First, functionally, DgPg needs to
create summaries. We draw inspiration from trans-
formers, especially because of its ability to produce
patterns that can be used as summaries. Mechanisti-
cally, our goal is to identify and present patterns that
can represent the shared practices. Our aim is not to
report all deviations from the shared practice nor to
evaluate or label a coding practice as good or bad.
Second, we aim for the DgPg model to be small
in size and fast to train. The compactness affects de-
ployment flexibility, but more importantly, affects the
time to train the model. We prefer the model to be
able to train quickly because, from what we have ob-
served, different healthcare providers do have highly
specialized and unique practices because of the kind
of care they provide to patients. We believe a smaller
and more specialized AI agent is easier to optimize,
control and audit. In principle, this has been proven in
the context of predicting an order sequence of charac-
ters in information theory (Grassberger, 2012), where
a distinction is made between algorithmic complexity
and statistical complexity. A sequence can be statis-
tically complicated but if it’s generated by a simple
algorithm, then its information can be quantified by
Table 1: An example of the ICD-10-CM code scheme being
inherently hierarchical.
code desc
S Injury, poisoning and certain other
consequences of external causes
S5
Injuries to the elbow and forearm
S52 Fracture of forearm
S52.5 Fracture of lower end of radius
S52.52 Torus fracture of ...
S52.521 ... of right radius
S52.521A Initial encounter for closed fracture
the complexity of the algorithm (Grassberger, 2012).
Even though the transformer architecture is new, we
believe the same principle still applies.
We aim to use the DgPg model in the following
way: given a starting point, e.g. a particular DX code,
we aim for DgPg to generate a graph of the most re-
lated DX codes that can be visualized as visual in-
sights to users. The formulation of our research prob-
lem is next.
3.2 Problem Formulation
A Unified Format. ICD-10-CM codes are inher-
ently hierarchical. Table 1 shows an example hierar-
chy, where one can trace an exact arm injury through
seven levels of details. The first level of the hierarchy
is encoded within the first character of the code, sec-
ond level in the second character, etc. DgPg needs an
architecture that can learn such coding schemes.
Typically, to use a transformer (Vaswani, 2017)
model requires one to restructure their problem at
hand as a sequence-to-sequence problem. Hence,
given our need for a model that learns the hierarchi-
cal coding schemes, the transformer architecture is a
natural choice. It allows our model to learn a code
piece-by-piece instead of all-at-once. In essence, we
will train our model to learn the first character, then
learn the next character based on the first character,
and iterate further recursively.
Baseline Memory Model. In contrast, let’s exam-
ine the problem of learning the relationship between
pairs of codes in a traditional way, where one has to
represent these codes as numeric indices. For exam-
ple, if the code A41.9 is the 123rd code out of 12,039
codes, then it would be encoded as the integer 123.
With this traditional formulation, a memory model
can be created as a matrix of 12,309× 12, 309. There
are two consequences: (1) the model is large (around
290 MB of float16 values), and (2) the model is not
generalizable.
Visualizing Medical Coding Practices Using Transformer Models
727
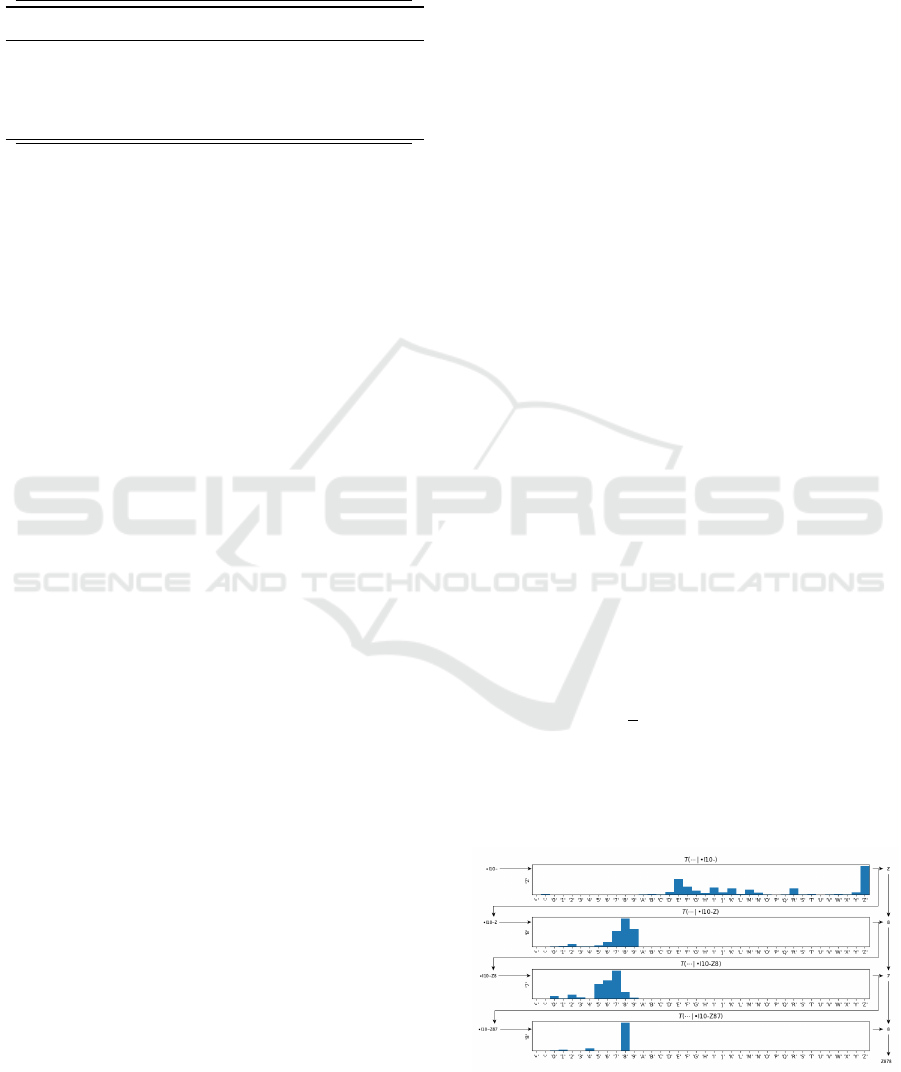
Table 2: Our data come from three datasets. In MIMIC-
IV, each record represents a single patient. For NHCS and
NHDS, a record can represent multiple patients. Hence, we
show the number of records and the number of patients rep-
resented by the datasets as two separate columns.
From Name # Records # Patients
MIT MIMIC-IV 430,812 430,812
CDC NHCS 123,565 33,595,765
CDC NHDS 756,953 111,514,788
Combined 1,277,400 145,507,435
As a specific example, consider a dataset where
60 % of the time the code A41.9 is followed by
I10.0; and 40 % of the time it is followed by I59.1.
Ideally, we want the model to learn that 100 % of the
time, it should predict I as the first character.
For this example, a memory model, will lose the
hierarchy information of the codes. Hence we choose
to build a unified text-to-text model on the character-
level, so that the model can learn the hierarchical cod-
ing schemes naturally as shown in Table 1.
Datasets. Our requirement for input data is mini-
mal. As we are only interested in the sequence of DX
codes, any dataset that has ordered sequences of DX
codes is sufficient. In this work, we use three datasets:
MIMIC-IV, NHDS, and NHCS. Table 2 details these
datasets.
The MIMIC-IV (Johnson et al., 2023) dataset is
an anonymized collection of medical records from
the Beth Israel Deaconess Medical Center. The data
spans multiple years. Within MIMIC-IV, we use the
hosp/diagnoses_icd data which has four columns:
admission number, sequence number, ICD version,
and ICD code.
The National Hospital Discharge (and Care) Sur-
veys (US CDC NCHS, 2010; US CDC NCHS, 2020)
cover different years: NHDS from 1996 to 2010, and
NHCS for only 2020.
These two CDC datasets follow a similar struc-
ture with 1 + N columns: a weight column w, and N
columns of DX codes. NHDS has N = 7 DX codes;
NHCS has N = 30 DX codes. We treat the column
ordering as the representative order of the codes, i.e.
DX1 is the first code, DX2 is the second, etc.
The weight column w serves as a frequency mul-
tiplier, to indicate one record in the dataset can repre-
sent multiple patients. For each input record, we emit
w copies as actual records for DgPg to learn.
Different datasets have different levels of speci-
ficity of the codes they use. In addition, to test the
generality of our data and our models, we choose a
common encoding scheme for all codes. Even though
MIMIC-IV has up to seven characters in the DX
codes and abides by ICD-10-CM standard, NHDS has
DX codes limited to four characters and are under
the ICD-9-CM standard. Hence we have converted
ICD-9-CM codes to ICD-10-CM codes, using the Na-
tional Bureau of Economic Research (NBER) ICD-
9/10 crosswalk. We’ve also truncated the length of
the DX codes in MIMIC-IV to four characters as well.
Every ICD-10-CM code is normalized to four charac-
ters, truncating and/or padding with hyphens as nec-
essary.
These data processing is to compare DgPg’s
model results across the MIMIC-IV dataset and the
two CDC datasets. DgPg’s capability is not limited to
four characters.
The DgPg Model: T (D). Notationally, we con-
sider T (D), where T is a DgPg transformer model
trained on some dataset D.
For a given model and dataset pair, we measure
the performance PERF using the standard approach
for transformer models: fixed-length sequence per-
plexity (Radford et al., 2019). To get an average
model performance, the PERF function gets applied
to 10 K samples from the validation split and are ag-
gregated via an arithmetic mean.
Specifically, we evaluate on sequences of codes.
For example, for a sequence of two codes,
(M542,G441), the DgPg model would see the se-
quence •M542G441. For each character in this se-
quence, the transformer model returns a probability
of that character occurring, p = (p
1
, ··· , p
9
). We
are only interested in the model’s predictive power,
so that PERF (x | T (D)) is calculated only on the 2nd
code.
log(PERF) = −
1
4
9
∑
i=5
logP (x
i
| x
1
, ··· , x
i−1
) (1)
We show experimentation results in Section 4.
T : A
n
× A → R
Figure 1: Starting from I10-, we predict the most likely
code to follow: Z878.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
728
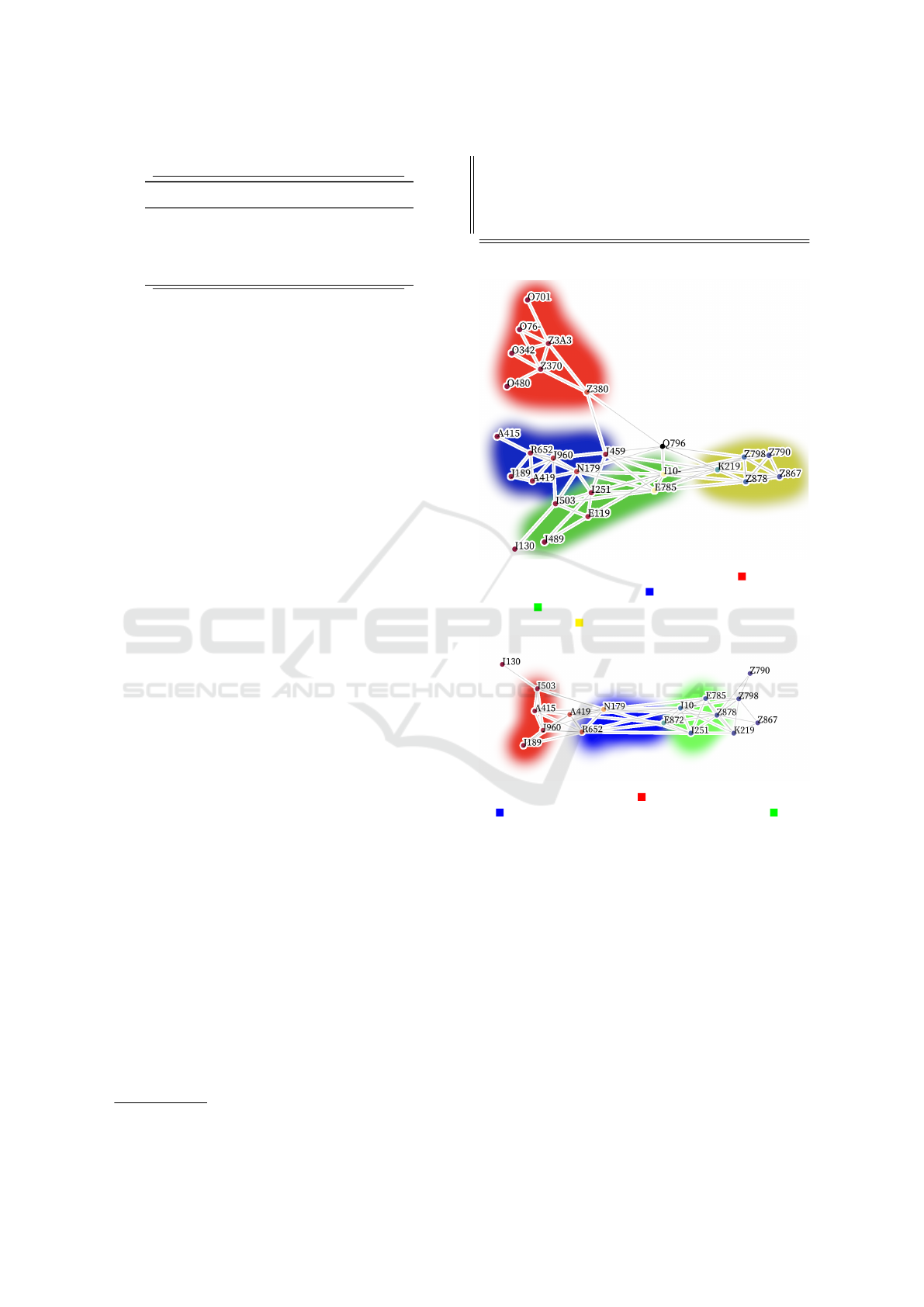
Table 3: An example of the data used to train our models.
Original Codes Training Sequence
DX1,DX2,DX3
N12,Q610,N179
A419,E119,""
•----N12-
•N12-Q610
•Q610N179
•N179----
T reports the probability that a character in an al-
phabet A follows a sequence of characters. T is in-
tended to be used iteratively: starting with a 4 charac-
ter code, predict the 1st character of the second code.
Then, starting with 4 + 1 characters, predict the 2nd
character of the second code. A complete example is
shown in Figure 1.
Table 3 is an example of the kind of input the
model is trained on.
4 RESULTS AND DISCUSSIONS
4.1 Configurations and Training
Base Model Configuration. We trained our mod-
els using a modified version of the nanoGPT project
1
.
We based our model configuration on nanoGPT’s in-
cluded configuration for character-level training on a
dataset of Shakespeare’s plays. This configuration for
our base model is in Listing 1. We also experimented
with different model sizes (Section 4.3).
Training Data. In Section 4.4, we trained on four
datasets: MIMIC, NHCS, NHDS datasets, and a 4th
dataset, which we refer to as “Combine”, which com-
bines all three of the MIMIC, NHCS and NHDS
datasets.
Compute Configuration and Training Time. All
of our models were trained on Google Colab using
a NVIDIA T4 GPU instance. On average, it took
1,092 S to train a model, at a cost of 0.16 USD/HR,
which amounts to to about 0.05 USD per model. As is
typical in transformer model training, DgPg uses half
precision f16 floating point numbers (Warner et al.,
2024).
Summary. When training the base model and the
variation models, the only technical difference is with
how the training data is arranged. In terms of train-
ing time, we notice very minor differences among the
DgPg base model and the variation models. Thus we
1
https://github.com/karpathy/nanoGPT
lea r n i n g _ r a t e = 1e -3 ; dr o p out = 0. 2
lr_decay_iters = 30 000 ; m in_ l r = 1e -4
be ta2 = 0. 99 ; ma x _ i t e rs = 30 000
ba t c h _ s i z e = 256 ; n_ l a yer = 3
n_ h ead = 3 ; n_ e mbd = 192
Listing 1: A summary of the base model configuration.
(a) Q79.6: Ehlers-Danlos Syndrome. Red is preg-
nancy and childbirth; Blue is other diverse diseases;
Green is circulatory, endocrine, and metabolic dis-
eases; Yellow is health status and personal history.
(b) A41.9: Sepsis. Red is acute conditions; Blue
is symptoms and chronic conditions; Green is
metabolic and circulatory conditions.
Figure 2: Visualization of diagnosis-specific coding pat-
terns (Section 4.2).
conclude that the cost to train for all DgPg models can
be regarded as the same. The reason to choose a par-
ticular DgPg model will solely depend on the use case
at hand.
4.2 Visualizing Coding Practices
We aim to visualize medical coding patterns using our
DgPg model. We designed a generative algorithm to
extract the most likely codes that follow or precede
a “seed code.” The generative approach to coding is
critical as it allows our models to learn and reproduce
the DX coding hierarchy. We then recursively conduct
Visualizing Medical Coding Practices Using Transformer Models
729
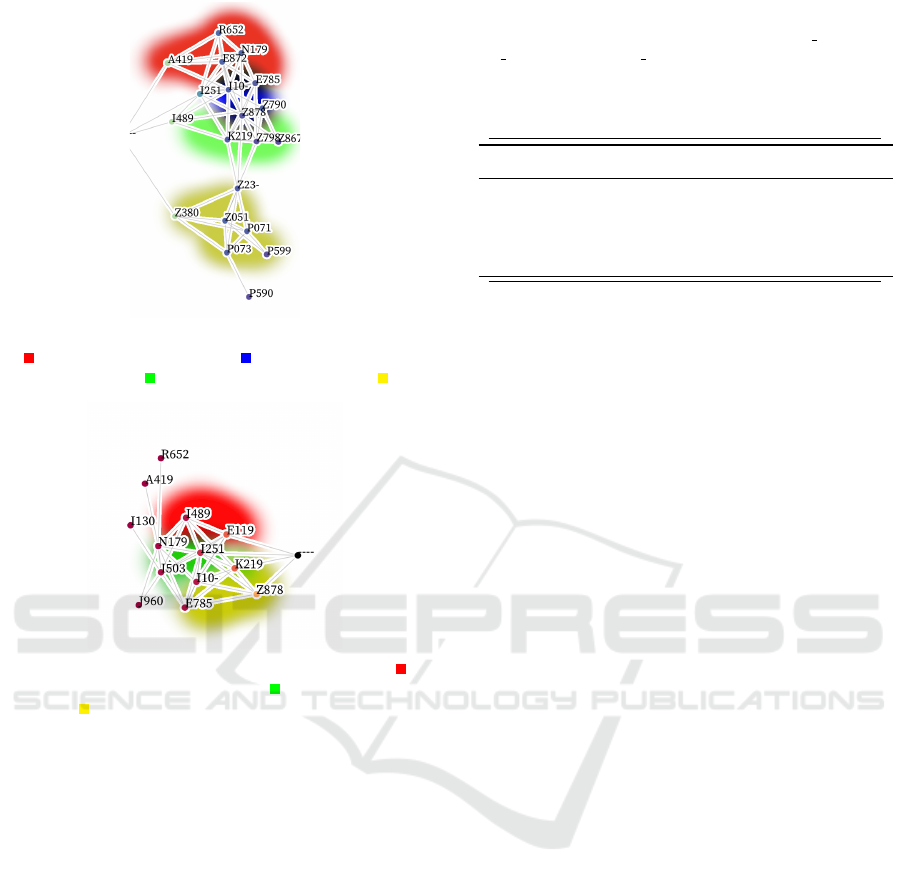
(a) The diagnoses most likely to be coded first. Red
is infectious diseases; Blue is circulatory system
diseases; Green is digestive diseases; Yellow is
health status and perinatal.
(b) The diagnoses most likely to be coded last. Red
is cardiovascular diseases; Green is comorbidities;
Yellow is factors influencing health status.
Figure 3: Visualization of which codes are most likely to be
coded (a) first and (b) last (Section 4.2).
a breadth-first search. For each code, we exhaustively
check the PERF of all following codes. The top k = 3
performers get added to the working set. This contin-
ues until we reach a depth of n = 3.
The resulting graphs (Figure 2 and 3) can be used
to understand the most common following and pre-
ceding codes for a given diagnosis.
One of those examples is in Figure 2a, which
details the codes most likely to surround a Ehlers-
Danlos diagnosis. Based on the graph, we can see
that the diagnosis is often preceded by records of the
patients birth, or the existence of other diseases. Our
graph also highlights a few codes that are used in sim-
ilar positions within the coding sequence, specifically
the codes that represent systemic conditions.
Figure 2b shows that sepsis is often preceded by
acute medical conditions that could have contributed
to a patient becoming septic. Similarly, it is often fol-
lowed by chronic, metabolic, and circulatory condi-
tions that sepsis could adversely affect.
Table 4: We evaluated the performance of the model at four
different levels of model complexities. NL is n layer, NH
is n head, and NE is n embd. The parameters in this table
override the base configuration in Listing 1. We found that
T
11 M
and T
1.3 M
perform similarly, despite a 10× difference
in model size.
Name NL NH NE Model Size Loss
T
11 M
6 6 384 20. MB 1.16
T
1.3 M
*
3 3 192 2.5 MB 1.17
T
230 K
2 2 96 440 KB 1.19
T
30 K
1 1 48 58 KB 1.25
Figure 3a (and 3b) visualize the most likely diag-
noses to be coded first (and last) in the diagnosis se-
quence. These graphs provide insight into what kinds
of codes are the most significant when starting or end-
ing the coding process.
The graphs we create can be of arbitrary size and
constraints. For Figure 2 and 3, each graph took
around 2 minutes to generate. Each graph is orga-
nized from left (earlier codes) to right (later codes).
4.3 Model Complexity
Here we report our ablation study to evaluate the ef-
fect of model configuration on the performance of
DgPg. Specifically, we experiment with the size of
the model. Because the tests are about model size, in
this section we refer to models in the test by the size of
the models, such as T
11 M
and T
1.3 M
, which represents
models with 11 M and 1.3 M parameters, respectively.
In the ablation study, we start with model config-
urations where we have no risk of overfitting the data.
We note that there are 12.3 K suitable (4-character
or less) diagnosis codes. Constructing a matrix of
transition probabilities would require 12.3 K ×12.3 K
float16 values, or around 290 MB. We limit our
model sizes to a maximum of 10 % of this size. This
limitation ensures that our models are learning gener-
alizable patterns.
Table 4 shows the results of our ablation tests. In
total, we tested four different model configurations.
From these results, we observe that T
11 M
and T
1.3 M
perform similarly even though their sizes differ by a
factor of 10×. With this result, we feel confident that
the T
1.3 M
configuration represents the optimal trade-
off between model size and performance. Results in
Section 4.2 uses T
1.3 M
.
We also note that even the T
230 K
configuration
could be suitable because its performance is compa-
rable to the other two best-performing models. T
30 K
,
however, is unsuitable as it is the worst performing
model.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
730
4.4 Discussion
Transformer Model Variations. We have also ex-
perimented with several variations on the base DgPg
model. Our base model can predict which DX codes
directly follow other DX codes within a medical
record. Related to but beyond this functionality, one
might also want the capability to predict codes that
precede other codes, and to expand the meaning of
“follows” to include “broadly follows a code” instead
of “immediately follows a code.” To this end, it’s use-
ful to build models that can predict broadly-following
codes can be used for normalizing the ordering of
codes within a medical record.
Furthermore, although a single model could be
used for all of these variations simultaneously via a
unique prompt prefix, our experiments show that spe-
cialized models can be built very efficiently. That is
because the DgPg framework is general, where the
main difference between each model variation lies
solely in the training data used. The format of the
training data stays the same while the meaning behind
each training sample varies. Due to this architectural
advantage, every model variation trains in the same
amount of time and using exactly the same DgPg
training pipeline. Results from the variation models
are not included in this publication due to the limit of
space, however.
Applicability of Transformers. The first reason we
chose the transformer architecture was because of the
need to learn the coding hierarchies. There is a sec-
ond reason as well. That is, generative models are
also a natural fit for generating summaries and, in this
paper, visual summaries, to explain large, messy real-
world datasets. As shown in Figure 2 and Figure 3,
the DgPg transformer model can perform well in such
a generative setting. We believe there is a strong po-
tential to extend DgPg’s organization-level analytics
use case into more clinically-oriented settings. From
this respect, DgPg is simply a starting point.
Limited Transferability. With the evidence that
patterns exist in all of our tested datasets, we also
want to know if they have the same patterns or if there
are unique patterns within each dataset. To evalu-
ate this, we leverage our PERF metric to measure a
model trained on one dataset when evaluated on an-
other dataset.
Using our DgPg model with 1.3 M parameters as
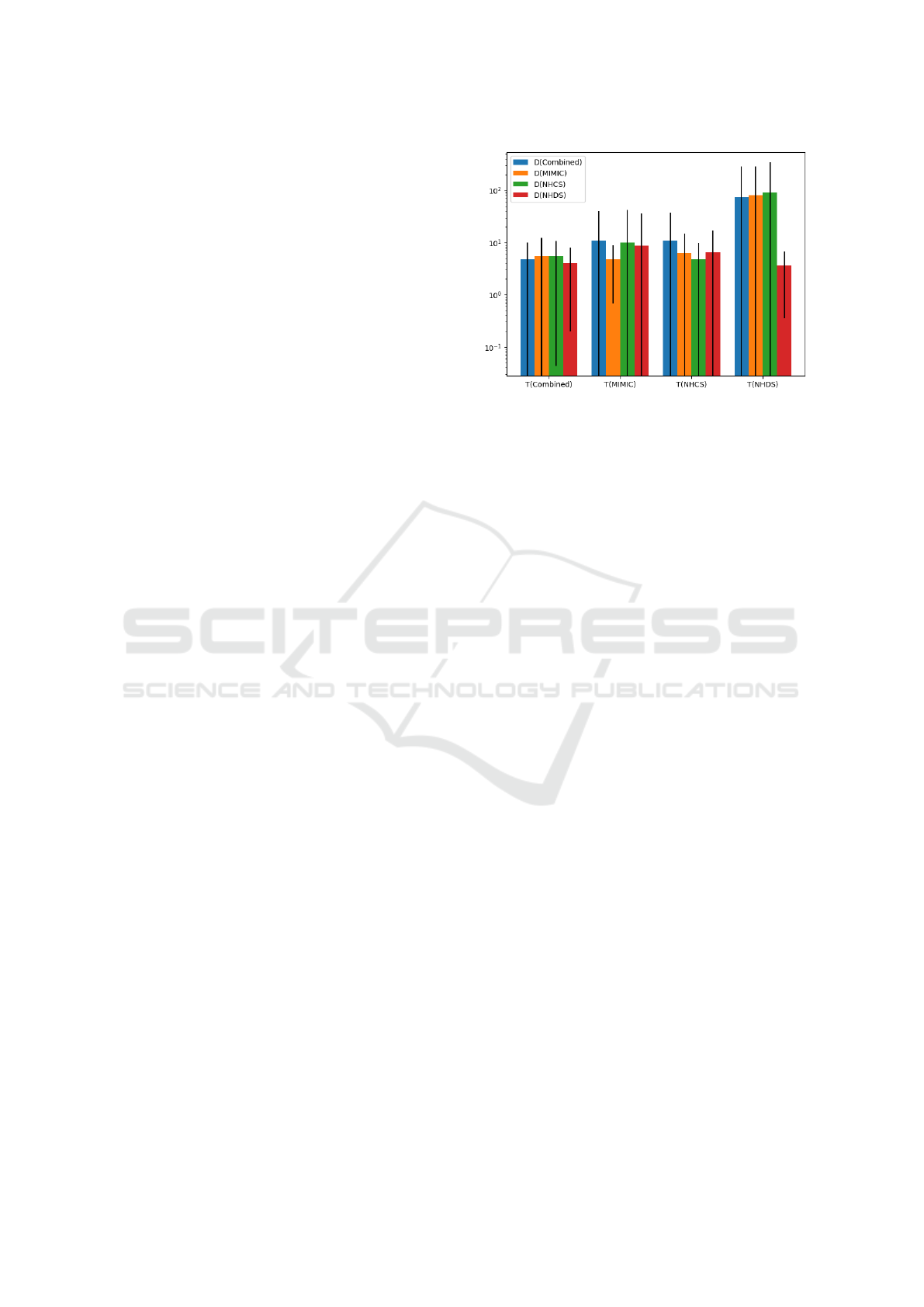
the example, Figure 4 shows how generalizable the
patterns are between datasets. There are two take-
aways from these results:
Figure 4: Evidence that the models have learned patterns
within their respective datasets that aren’t present in the
other datasets. T (·) represents that the model was trained
on a dataset, D(·) represents the dataset that was tested. For
each dataset, 10 K samples were collected. For each sample,
the model’s performance PERF was calculated. The mean
and standard deviation of the 10 K samples is shown in this
chart. As expected, testing with the same dataset that was
trained on yields a better PERF than testing on a different
dataset than the model was trained on. MIMIC and NHCS
are the most generalizable models, while NHDS is the least
generalizable.
First, as expected, the model trained on the Com-
bined data performs equally well on all of the other
datasets that make it up. This reaffirms the gener-
ally held belief that training on an increased variety
of datasets yields improved performance.
Second, the model trained only on NHDS data
performs the worst when tested on all the other
datasets. This is evidence that the patterns within
NHDS are unique to that dataset and are less general-
izable to other medical records datasets.
5 CONCLUSION
In this paper, we developed a transformer model,
DgPg, that can learn organization-level analytics and
summarize representative coding patterns based on
any corpus of medical records. Training a DgPg
model for a medical records dataset is both time- and
cost-efficient. After training, the DgPg models are
compact in size. We also demonstrate the use of DgPg
models in a generative use case to visualize the rep-
resentative coding patterns, as well as in a classifica-
tion use case to evaluate how representative a code
sequence is, given the context of any given dataset.
This work is only an initial step showing that
transformer models can learn the representation of
coding patterns. As future work, we aim to investigate
possibilities to continuously improve the capabilities
Visualizing Medical Coding Practices Using Transformer Models
731

of DgPg and to use transformer models in a bigger
variety of clinical use cases. In addition, as halluci-
nation is a research priority for language models, we
want to also study the issue of hallucination as it’s ap-
plicable to DgPg transformer models too.
REFERENCES
Abr
`
amoff, M. D., Lavin, P. T., Birch, M., Shah, N., and
Folk, J. C. (2018). Pivotal trial of an autonomous
ai-based diagnostic system for detection of diabetic
retinopathy in primary care offices. NPJ digital
medicine, 1(1):39.
Bai, T. and Vucetic, S. (2019). Improving medical code
prediction from clinical text via incorporating online
knowledge sources. In The World Wide Web Confer-
ence, pages 72–82.
Brown, T. B. (2020). Language models are few-shot learn-
ers. arXiv preprint arXiv:2005.14165.
Chanda, A. K., Bai, T., Yang, Z., and Vucetic, S.
(2022). Improving medical term embeddings using
umls metathesaurus. BMC Medical Informatics and
Decision Making, 22(1):114.
CMS (2024). ICD-10-CM Official Guidelines for Cod-
ing and Reporting FY 2025. United States CMS
https://www.cms.gov/medicare/coding-billing/icd-10-
codes.
Devlin, J. (2018). Bert: Pre-training of deep bidirec-
tional transformers for language understanding. arXiv
preprint arXiv:1810.04805.
Grassberger, P. (2012). Randomness, information, and com-
plexity. arXiv preprint arXiv:1208.3459.
Henry, K. E., Hager, D. N., Pronovost, P. J., and Saria,
S. (2015). A targeted real-time early warning score
(trewscore) for septic shock. Science translational
medicine, 7(299):299ra122–299ra122.
Hsu, C.-C., Karnwal, S., Mullainathan, S., Obermeyer,
Z., and Tan, C. (2020). Characterizing the value
of information in medical notes. arXiv preprint
arXiv:2010.03574.
Hu, S., Teng, F., Huang, L., Yan, J., and Zhang, H. (2021).
An explainable cnn approach for medical codes pre-
diction from clinical text. BMC Medical Informatics
and Decision Making, 21:1–12.
Johnson, A. E., Bulgarelli, L., Shen, L., Gayles, A., Sham-
mout, A., Horng, S., Pollard, T. J., Hao, S., Moody,
B., Gow, B., et al. (2023). Mimic-iv, a freely acces-
sible electronic health record dataset. Scientific data,
10(1):1.
Kim, B.-H. and Ganapathi, V. (2021). Read, attend, and
code: Pushing the limits of medical codes prediction
from clinical notes by machines. In Machine Learning
for Healthcare Conference, pages 196–208. PMLR.
Koleck, T. A., Tatonetti, N. P., Bakken, S., Mitha, S., Hen-
derson, M. M., George, M., Miaskowski, C., Smal-
done, A., and Topaz, M. (2021). Identifying symptom
information in clinical notes using natural language
processing. Nursing research, 70(3):173–183.
Krittanawong, C., Zhang, H., Wang, Z., Aydar, M., and Ki-
tai, T. (2017). Artificial intelligence in precision car-
diovascular medicine. Journal of the American Col-
lege of Cardiology, 69(21):2657–2664.
Malhotra, K., Hobson, T. C., Valkova, S., Pullum, L. L., and
Ramanathan, A. (2015). Sequential pattern mining of
electronic healthcare reimbursement claims: Experi-
ences and challenges in uncovering how patients are
treated by physicians. In 2015 IEEE International
Conference on Big Data (Big Data), pages 2670–
2679. IEEE.
Mullenbach, J., Wiegreffe, S., Duke, J., Sun, J., and
Eisenstein, J. (2018). Explainable prediction of
medical codes from clinical text. arXiv preprint
arXiv:1802.05695.
Radford, A., Wu, J., Child, R., Luan, D., Amodei, D.,
Sutskever, I., et al. (2019). Language models are un-
supervised multitask learners. OpenAI blog, 1(8):9.
Raffel, C., Shazeer, N., Roberts, A., Lee, K., Narang, S.,
Matena, M., Zhou, Y., Li, W., and Liu, P. J. (2020).
Exploring the limits of transfer learning with a unified
text-to-text transformer. Journal of machine learning
research, 21(140):1–67.
Raji, M., Duggan, J., DeCotes, B., Huang, J., and Van-
der Zanden, B. (2018). Modeling and visualizing stu-
dent flow. IEEE Transactions on Big Data, 7(3):510–
523.
US CDC NCHS (2010). National Hospital Discharge Sur-
vey (NHDS). US Department of Health and Human
Services, Centers for Disease Control.
US CDC NCHS (2020). National hospital care survey
(NHCS). CDC Stacks.
Vaswani, A. (2017). Attention is all you need. Advances in
Neural Information Processing Systems.
Warner, B., Chaffin, A., Clavi
´
e, B., Weller, O., Hallstr
¨
om,
O., Taghadouini, S., Gallagher, A., Biswas, R., Lad-
hak, F., Aarsen, T., et al. (2024). Smarter, better,
faster, longer: A modern bidirectional encoder for
fast, memory efficient, and long context finetuning
and inference. arXiv preprint arXiv:2412.13663.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
732
