
Reconstruction of 3D Brain Structures from Clinical 2D MRI Data
Rui Shi
1 a
, Tsukasa Koike
2 b
, Tetsuro Sekine
3 c
, Akio Morita
4 d
and Tetsuya Sakai
1 e
1
Department of Computer Science and Engineering, Waseda University, Shinjuku, Tokyo, Japan
2
Department of Neurosurgery, Teraoka Memorial Hospital, Fukuyama, Hiroshima, Japan
3
Department of Radiology, Nippon Medical School Musashi Kosugi Hospital, Kawasaki, Kanagawa, Japan
4
Tokyo Rosai Hospital, Ota, Tokyo, Japan
Keywords:
Brain MRI, 3D Reconstruction.
Abstract:
As the population is aging worldwide, the number of dementia patients increases. Brain MRI is expected to
play a crucial role in the prediction of dementia at an early stage. Current 3D brain structure reconstruction
methods have strict rules and require a large number of slice images. Routine clinical MRI files contain much
fewer slices, but diagnosis relies heavily on information obtained from MRI scans. In this paper, we proposed
a method that is able to reconstruct the 3D brain structure with 2D DICOM MRI images within the clinical
routine budget, by applying trilinear interpolation. The generated images and structures are evaluated with
PSNR and SSIM. The results show that although the details in the generated 2D slices are not ideal, our
method is able to reconstruct 3D structures that are highly similar to the original brain structures using only
one-fifth of the image slices.
1 INTRODUCTION
Population around the world is aging. For example,
in Japan, the proportion of people aged 65 and over
exceeds one-quarter of the total population (Statis-
tics Bureau, 2023). Providing proper medical care for
senior citizens is a major issue and needs to be ad-
dressed with contributions from society.
Dementia is one of the most common mental ill-
nesses that afflict the elderly, which causes a decline
in cognitive abilities and has negative impacts on the
individual in various ways, such as mobility, emotion,
and social relationships (van der Flier and Scheltens,
2005). Since dementia is slowly progressive (Bathini
et al., 2019), if diagnosed at the early stage, the devel-
opment of the disease can be eased with intervention.
Such prediction models are expected to be developed
with Brain MRI images.
Magnetic resonance imaging (MRI) technology
has been used for decades to obtain high-quality brain
images. Compared to other diagnostic imaging tech-
niques, MRI does not expose patients to radiation
in Positron Emission Tomography (PET) (Carlson
a
https://orcid.org/0009-0002-7339-0305
b
https://orcid.org/0000-0002-9931-0716
c
https://orcid.org/0000-0003-1547-6696
d
https://orcid.org/0000-0002-2497-5772
e
https://orcid.org/0000-0002-6720-963X
and Carlson, 2007), and provides better tissue con-
trast than Computed Tomography (CT) Scanner (Ebel
and Benz-Bohm, 1999). Various information can be
obtained through Brain MRI, such as the changes
in white matter which reflect systemic hypertension
(Salerno et al., 1992).
MRI scans divide the brain into slices at given in-
tervals, where each slice shows a 2D scan image of
the corresponding location. During the slicing pro-
cess, 3D characteristics of the patient’s brain can be
lost. The volume and proportions become difficult to
quantify, and the spatial relationships between differ-
ent parts are unclear. But most current diagnoses and
surgery plans are based on MRI scans. Hence, it is
essential to reconstruct the 3D structure of the brain
for accurate presentation and alignment.
DICOM is the standard format used in hospitals
and clinics for MRI (Mustra et al., 2008). If the 3D
brain structure is reconstructed by simply stacking the
slice images according to their location, at least 100
slices are required for each coordinate axis, or a total
of more than 300. However, in clinical routine, DI-
COM MRI files are usually generated with only 19
slices along one single coordinate axis for financial
and waiting time considerations. Diagnoses based on
such files highly depend on the expertise and experi-
ence of the doctor.
To fill the gap and better assist doctors in diagno-
sis and surgery decisions, in this work, we propose a
352
Shi, R., Koike, T., Sekine, T., Morita, A. and Sakai, T.
Reconstruction of 3D Brain Structures from Clinical 2D MRI Data.
DOI: 10.5220/0013259000003905
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 14th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2025), pages 352-359
ISBN: 978-989-758-730-6; ISSN: 2184-4313
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

method to reconstruct the 3D brain structure with 2D
MRI images at a relatively low cost, with a limited
number of slices within the clinical routine budget,
while aiming for high quality, accuracy, and speed.
2 PRELIMINARIES
2.1 Background
2.1.1 Metadata in DICOM Files
Each DICOM file has a metadata header, providing
information such as slice spacing and dimension. To
achieve success in 3D reconstruction by stacking the
2D slices, the required number of slices exceeds what
is obtained in most routine clinical MRI DICOM files,
and the metadata of the slices must match in a highly
accurate order (AlZu’bi et al., 2020). However, vibra-
tion during the scan process and variations between
acquisition devices can lead to divergences and inac-
curacies in the metadata.
In addition, routine clinical MRI slices are mostly
separated with relatively large spacing, leaving the
structures between the intervals vacant. By simply
stacking the images, the generated structure is discon-
tinuous because of the information lost in the inter-
vals, and differs largely from the actual structure.
2.1.2 Bias Field and Noise
Bias field exists during MRI scans, especially those
taken by old MRI machines. Such uneven intensity
produces smooth, low-frequency signals that interfere
with the MRI scanning process (Juntu et al., 2005).
Bias field can cause intensity variation among homo-
geneous tissues, which changes high-frequency com-
ponents such as contours and edges, causing blurs in
the images. The variation may not be easily observed
but can result in the decline of feature points during
image processing.
Also, the RF emission from the thermal mo-
tion of the patient’s body causes much inevitable
noise in the scanned MRI images, which appears
as irregular granular patterns and confuses informa-
tion acquisition (Aja-Fern
´
andez and Vegas-S
´
anchez-
Ferrero, 2016).
2.2 Related Work
2.2.1 Pre-Processing of Image
Histogram equalization is often used to counteract the
effects of bias fields (Senthilkumaran and Thimmi-
araja, 2014). It is a general method used for grayscale
image enhancement, performed with the cumulative
distribution function (CDF) of intensity. Since image
voxels in DICOM files are encoded by intensity, they
resemble the grayscale images, which share the same
color channel. The images processed by histogram
equalization will have higher contrast than the origi-
nal images.
Noise filters are also widely used to improve the
quality of scanned images. There are various types of
noise filters, realized with different approaches. For
example, in the work of Thanh and Hai (Thanh and
Hai, 2017), a mean-unsharp filter, which is convo-
luted with the 2D MRI images, is applied to remove
the noise.
2.2.2 Marching Cube
Marching cubes is a classic computer graphics algo-
rithm that can produce high-resolution 3D surfaces
with simple construction operations (Lorensen and
Cline, 1998). Even in recent years, this algorithm has
shown good performance in reconstructing 3D knee
surfaces and spine structures from 2D CT images (Pa-
tel and Mehta, 2012), but the calculation will be slow
if a large amount of 2D data is processed.
2.2.3 Interpolation
Interpolation is a mathematical approach to estimate
unknown points based on the existing points. By con-
structing a continuous function that passes through
the given data points, unknown points between the
given data points can be approximated with a high ac-
curacy (Davis, 1975). In Ghoshal’s work (Ghoshal
et al., 2020), highly accurate 3D reconstruction of
spine is achieved by combining the Marching cubes
algorithm and interpolation. Although not quantified,
reconstruction works of 3D brain structure from 2D
MRI images have also received satisfying results us-
ing trilinear interpolation (Fajar et al., 2022) (Thanh
and Hai, 2017).
3 METHOD
In this section, we propose a method that is able to re-
construct the 3D brain structure from a small amount
of 2D DICOM MRI images along one single coordi-
nate axis. Figure 1 shows the four steps taken to ac-
complish the process, which are introduced in detail.
3.1 Pre-Processing of Image
We propose two alternative approaches for contrast
enhancement and noise cancellation. Approach A in-
Reconstruction of 3D Brain Structures from Clinical 2D MRI Data
353
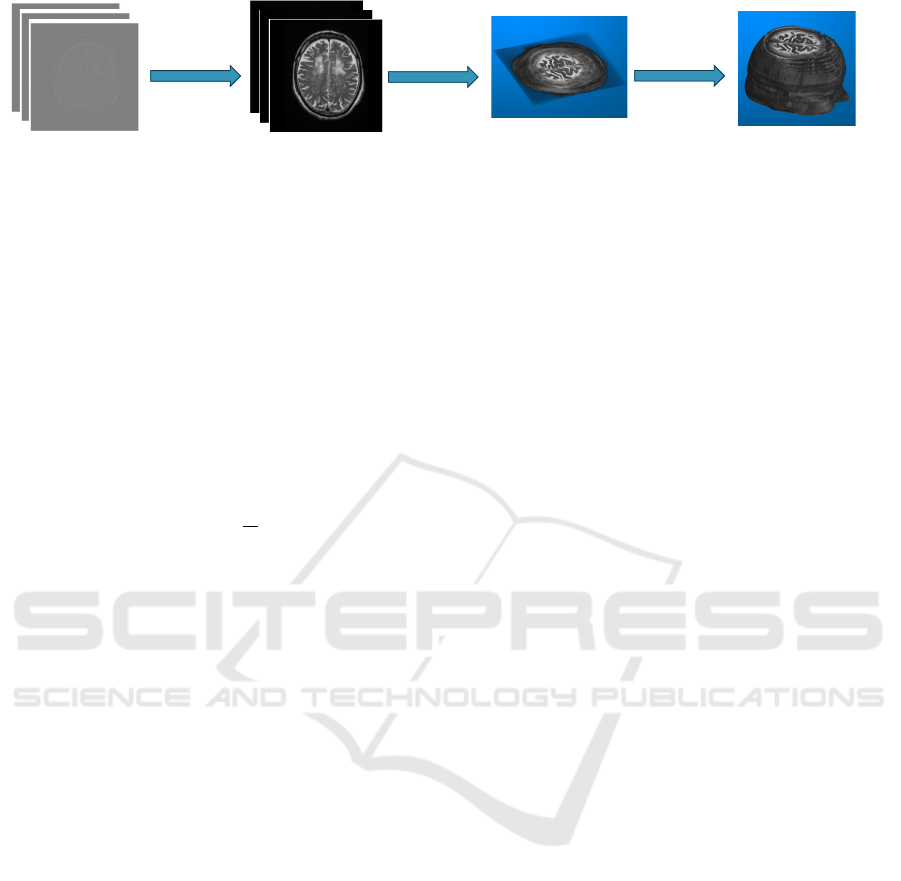
Original image slices
T2 (Axial)
- Case 1 2007/03/30 from the Test Dataset
Image slices after Pre-processing
Resized Reconstructed 3D structure
with transparent background
Step 1
Step 2
Primitive Reconstructed 3D
Structure
Step 3 & 4
Figure 1: Workflow of 3D Brain Reconstruction from Clinical 2D MRI Image data. 1) Pre-processing of the image; 2)
Reconstruction of 3D structure with Trilinear Interpolation; 3) Resizing the generated 3D Image; 4) Making the Background
Transparent. The demonstrated original images are from the MRI record collected on Patient Case 1 from the Test Dataset on
March 30, 2007. Further information on the Test Dataset can be found in Section 4.
volves histogram equalization and a noise filter, while
Approach B comprises only one contrast filter.
3.1.1 Histogram Equalization
Histogram equalization is applied to enhance the con-
trast in the images (Fajar et al., 2022). Considering
in a discrete grayscale image x, the probability of the
occurrence of a pixel of level i in the image is:
p
x
(i) = p(x = i) =
n
i
n
, 0 ≤ i < L (1)
where L is the maximum pixel value, n
i
is the
number of occurrences of the pixel of level i, and n
is the number of pixels in the image.
As an MRI DICOM image is similar to a grayscale
image with intensity ranging from 0 to 255 for every
voxel, L equals the intensity level bound 256. The
CDF for the intensity v can be calculated as
cdf
x
(v) =
v
∑
j=0
p
x
(x = j) (2)
The histogram-equalized intensity value h(v) is
derived as
h(v) = round(cdf
x
(v) · (L − 1)) (3)
Through histogram equalization, the equalized in-
tensity value replaces the original value, distribut-
ing the pixels more evenly throughout the full range,
which gives the image a higher contrast.
3.1.2 Noise Filter
Our noise filter is developed with homogenization of
the background.
First, we verify the boundary of the skull. As the
objects within the head are separated from the back-
ground by the skull, which preserves a much brighter
shade than the surrounding background, we can eas-
ily measure the intensity value of the skull and set the
value as a threshold.
Once the threshold is obtained, the head boundary
can be detected. The noise or irrelevant background
can be effectively removed by setting all pixels out-
side the identified boundaries to zero.
3.1.3 Contrast Filter
An alternative way to enhance the contrast and cancel
the noise in one step is to use a contrast filter. We de-
signed a contrast filter that adjusts the image contrast
by normalizing the pixel values to the full 8-bit range
0 to 255.
First, we find the maximum and minimum pixel
values in the image, and calculate the range of vari-
ance in pixel value by subtracting the minimum value
from the maximum value.
Then, we subtract the minimum pixel value from
each pixel and divide the subtraction results by the
range of pixel value variance to normalize the pixel
values to 0 to 1.
Finally, we multiply the division results by 255 to
scale the normalized values into 0 to 255.
Through the contrast filter, the pixels are dis-
tributed more evenly to the full range, and the abrupt
variance caused by noise is also mostly reduced, can-
celing noise during the normalization process.
3.2 Reconstruction of 3D Structure with
Trilinear Interpolation
After pre-processing, the 2D image slices are now
ready to be primitively combined to form the 3D
structure.
First, the 2D image slices are stacked according
to their slice location shown in the metadata. When
image data are simply stacked, there is much empty
space with missing information at the intervals be-
tween the slices. As the intervals are large for routine
clinical MRI images, the spacing may vary between
slices.
These empty intervals can be filled with the slices
generated by trilinear interpolation, which recon-
structs the 3D information between the slices and
smoothens the structure (Fajar et al., 2022). The aim
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
354
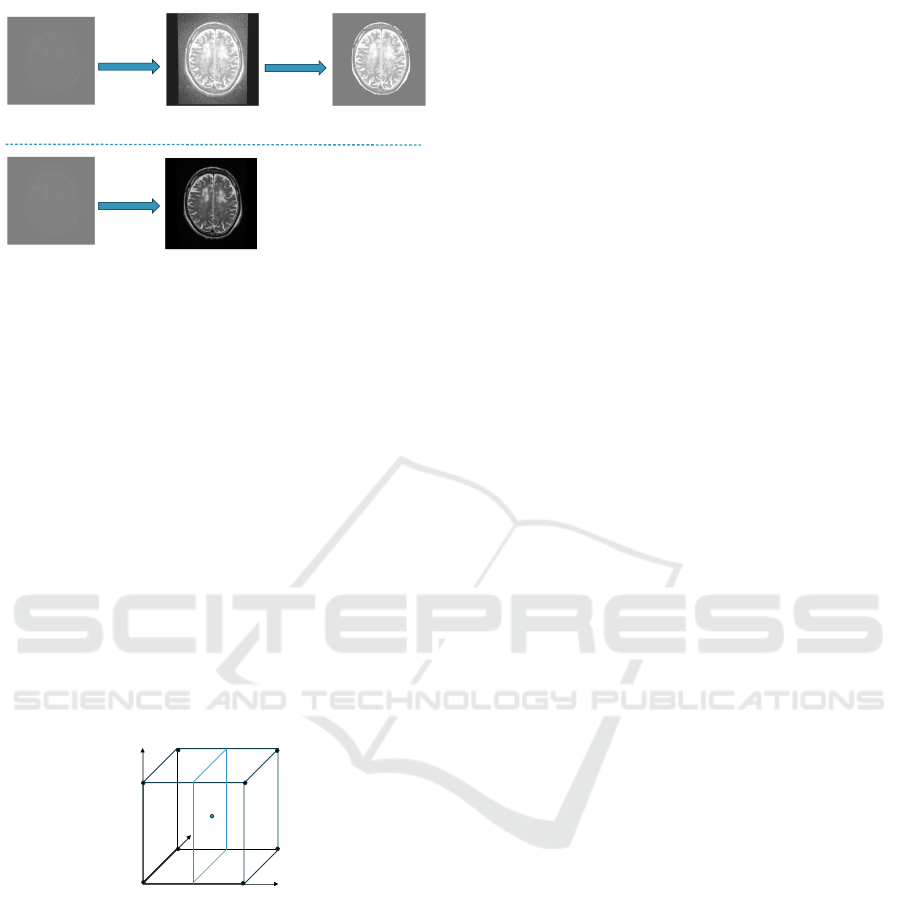
Image after histogram
equalization
Denoised image after
histogram equalization
Image after our
contrast filter
Original image
slices
T2 (Axial)
- Case 1 2007/03/30
Histogram
equalization
Noise Filter
Contrast
filter
Approach A
Approach B
Original image
slices
T2 (Axial)
- Case 1 2007/03/30
Figure 2: Workflows of the Alternative Approaches (Ap-
proach A and Approach B) for Contrast Enhancement and
Noise Cancellation. Approach A first applies Histogram
Equalization to the original image, then passes the equal-
ized image through the Noise Filter. Approach B only
passes the original image through the Contrast Filter. The
demonstrated original image is one slice from the MRI
record collected on Patient Case 1 from the Test Dataset
on March 30, 2007.
is to approximate the original 3D structures with our
interpolated structures.
Trilinear interpolation is a method of multivariate
interpolation on a 3D regular grid (Davis, 1975). It
can predict the pixel value of any point within the 3D
structure with a faster speed than the Marching cube
(Rajon and Bolch, 2003), as follows:
Considering in a unit-size cubic lattice, a point lo-
cated at (u, v, w) in the spatial domain expects pixel
prediction by trilinear interpolation, as shown in 3.
y
x
z
(u,v,w)
C1
C0
C3
C5
C2
C7
C6
Figure 3: An example of Trilinear Interpolation within a
unit-size cubic lattice. The 3D Cartesian coordinate system
is formed by the x, y, and z axes. The C
n
values are the pixel
values of the vertices, where n is the index for each vertex.
The point located at (u, v, w) is expecting pixel prediction.
The pixel value C(u, v, w) of the point located at
(u, v, w) is calculated as:
C(u, v, w)
=C
0
(1 − u)(1 − v)(1 − w) +C
1
u(1 − v)(1 − w)
+C
2
uv(1 − w) +C
3
(1 − u)v(1 − w)
+C
4
(1 − u)(1 − v)w +C
5
u(1 − v)w
+C
6
uvw +C
7
(1 − u)vw
(4)
3.3 Resizing 3D Image
The default setting makes the primitive reconstructed
3D structure a plate. Therefore, resizing is needed
to adjust the structure to the proper scale. We adjust
the proportions of the head referencing biostatistics
data (Bail et al., 2009) and uniformize the size and
resolution of the generated 3D brain structures.
3.4 Making the Background
Transparent
During the pre-processing step, we evenly enhanced
the image contrast, where the pixels representing the
objects within the head preserved a much different
intensity level from the pixels representing the back-
ground. We can set the pixels within the background
level to 0 to make the background transparent eas-
ily and efficiently, so that the brain structure can be
clearly viewed from all angles and is emphasized.
4 EXPERIMENT
In this section, the details of our experiments are clar-
ified.
4.1 Dataset
In this research, we use a Test Dataset and an Eval-
uation Dataset. Both datasets are collected from the
Teraoka Memorial Hospital in Hiroshima, Japan.
4.1.1 Test Dataset
The Test Dataset consists of 115 records of 2D MRI
DICOM files. Nineteen volunteers have contributed
to this dataset through the years.
Each 2D MRI DICOM file in this dataset includes
19 slices at a spacing interval of 5 mm. There are
three types of data scanned along the coordinate axes:
(1) The Axial data is collected along the vertical axis,
from the top to the bottom of the head; (2) The Coro-
nal data is collected along one horizontal axis, from
the back to the front of the head; (3) The Sagittal data
is collected along the other horizontal axis, orthogo-
nal to the Coronal axis, from the side of the head.
This dataset is used to verify whether our method
is adaptable to all kinds of data, as there is a relatively
rich number of samples and a variety of data types in
this dataset.
Reconstruction of 3D Brain Structures from Clinical 2D MRI Data
355

4.1.2 Evaluation Dataset
The Evaluation Dataset consists of 22 records of 3D
MRI DICOM files for evaluation. This dataset is con-
tributed by twenty-two volunteers.
In this dataset, there are 22 records of 3D Sagittal
data. Each record of such data includes approximately
100 slices, spaced with an interval of 1 mm.
This dataset is used to evaluate the generation per-
formance quantitatively. With this dataset, by taking
samples from the 3D Sagittal data, we can not only
obtain the similarity of the entire structure between
the original form and the generated form, but also the
similarity between the generated slice and the origi-
nal slice. In addition, if we take the first slice in every
five slices as a sample, we can create a sampled 2D
set containing approximately 20 slices, similar to the
routine clinical file containing 19 slices. Hence, we
can expect similar performance in reconstructing 3D
brain structure using the proposed method, with rou-
tine clinical MRI data.
4.2 Evaluation Metrics
Two evaluation metrics are applied to validate the re-
sults.
4.2.1 Peak Signal to Noise Ratio (PSNR)
Peak signal-to-noise ratio (PSNR) is an image eval-
uation measure. It is often used to calculate the vi-
sual error between two images, such as measuring the
visual difference between the original image and the
compressed image, the difference between the image
generated by the generative network and the actual
image, etc.
PSNR is defined via the mean squared error
(MSE). Suppose there are two m×n monochrome im-
ages I and K, I is noiseless and K is similar to I but
contains noise. MSE between I and K is defined as
MSE(I, K) =
1
mn
m−1
∑
i=0
n−1
∑
j=0
[I(i, j) − K(i, j)]
2
(5)
where I(i, j) and K(i, j) are the pixel values at posi-
tion (i, j) (i-th row and j-th column) in I and K re-
spectively.
PSNR is defined in units of dB, as
PSNR(I, K) = 10 · log
10
(MAX
I
)
2
MSE
(6)
where MAX
I
is the maximum pixel value of the orig-
inal image.
PSNR is always non-negative and equals infinity
when the two images are identical (Bull and Zhang,
2014). The higher the PSNR value, the less noise in
the noisy image. PSNR is a pixel-wise metric: if the
pixel value is different, it will be considered as noise
and quantified.
4.2.2 Structural Similarity Index Measure
(SSIM)
Structural similarity index measure (SSIM) is an-
other metric for measuring the similarity between
two images. SSIM performs the comparison from
a structural perspective, considering brightness, con-
trast, and structural information in the images (Wang
et al., 2004). Structural information assumes that pix-
els have strong inter-dependencies, especially when
spatially close.
Suppose there are two images x and y, SSIM be-
tween x and y is defined as
SSIM(x, y) = [l(x, y)]
α
[c(x, y)]
β
[s(x, y)]
γ
(7)
where
l(x, y) =
2µ
x
µ
y
+C
1
(µ
x
)
2
+(µ
y
)
2
+C
1
compares the brightness
between x and y;
c(x, y) =
2σ
x
σ
y
+C
2
(σ
x
)
2
+(σ
y
)
2
+C
2
compares the contrast be-
tween x and y;
s(x, y) =
σ
xy
+C
3
σ
x
σ
y
+C
3
compares the structure similarity
between x and y;
α, β, γ are the positive parameters for adjustment;
µ
x
, µ
y
are the pixel sample means of x and y;
σ
x
, σ
y
are the standard deviations of x and y;
σ
xy
is the covariance of x and y;
C
1
,C
2
,C
3
are constants to stabilize the division
with a weak denominator.
The higher the SSIM value is, the higher the sim-
ilarity the two images preserve. The maximum value
of SSIM is achieved when the two images are iden-
tical, and the SSIM value can be negative when the
structural information in the two images differs too
much.
5 RESULTS
In Section 2, we proposed two alternative approaches
for pre-processing. Figure 4 shows the processed im-
ages for a record with Approaches A and B, respec-
tively.
Both the original and the generated 3D brain struc-
ture can be presented and observed from all angles af-
ter the execution of the implemented program. Figure
5 shows a sample of the original and the generated 3D
brain structures from the same record.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
356
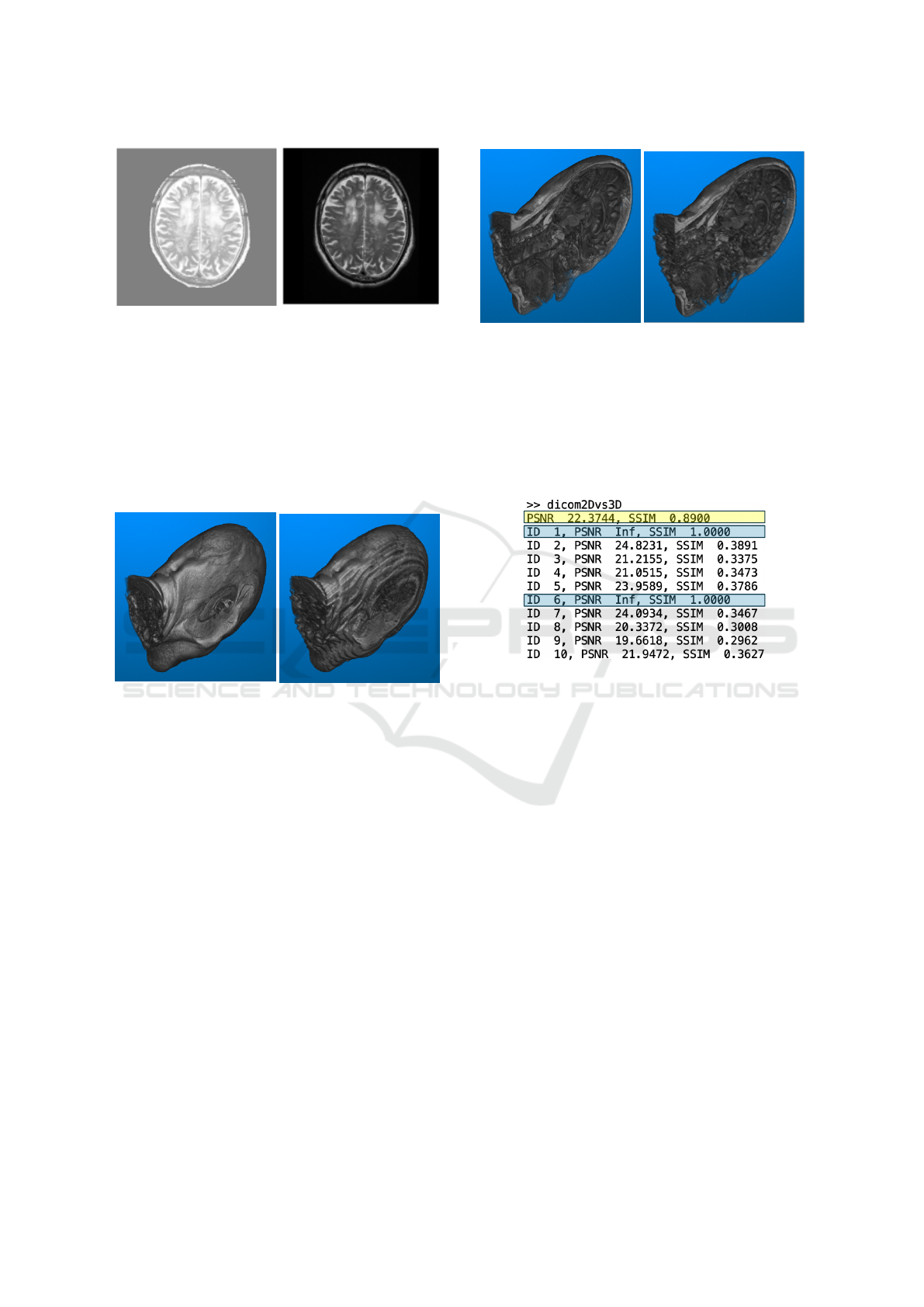
(a) Denoised image after his-
togram equalization.
(b) Image after our contrast
filter.
Figure 4: The 2D MRI images pre-processed with alterna-
tive approaches for Contrast Enhancement and Noise Can-
cellation. (a) Approach A, first applying Histogram Equal-
ization to the original image, and then passing the equalized
image through the Noise Filter. (b) Approach B, which only
passes the original image through the Contrast Filter. The
demonstrated images came from the same original image,
one slice from the MRI record collected on Patient Case 1
from the Test Dataset on March 30, 2007.
(a) Original structure.
(b) Generated structure.
Figure 5: An example of the Original and the Generated
3D Brain Structures from the Same Record. The left di-
agram shows the original 3D brain structure stacked with
103 slices along the corresponding axis. The right diagram
shows the interpolated 3D brain structure generated with 21
sampled slices along the same axis. The image data used by
both diagrams came from the same MRI record collected on
Patient Case 01 from the Evaluation Dataset.
Cross-sections can also be cut at arbitrary posi-
tions in the original and the generated 3D brain struc-
ture, allowing observation from all angles. Figure 6
shows a sample of the original and the generated 2D
brain slices from the same record.
Calculation results of the PSNR and SSIM val-
ues are printed out after execution of the implemented
program. Figure 7 presents an example showing part
of the output.
The average PSNR and SSIM results for each
patient are summarized in Table 1. The PSNR and
SSIM (Structure) values are obtained by comparing
the original and the generated 3D brain structures.
The SSIM (Slices) values are obtained by comparing
slice by slice.
(a) Original structure. (b) Generated structure.
Figure 6: An example of the Original and the Generated 2D
Brain Slices from the Same Record. The left diagram shows
a cross-section in the original 3D brain structure, while the
right diagram shows the cross-section at the same position,
from the same angle, in the interpolated 3D brain structure.
The image data used by both diagrams came from the same
MRI record collected on Patient Case 01 from the Evalua-
tion Dataset.
Figure 7: An example showing partial output. The values in
the yellow-marked area are obtained by comparing the orig-
inal and the generated 3D brain structures as an overview.
The rest of the output with IDs are obtained by making the
comparison slice by slice, between the original slices and
the corresponding slices at the same position from the inter-
polated 3D brain structure. The values in the blue-marked
areas are the results of the sampled slices. These results are
excluded from the statistical analysis later on. This output is
generated based on the data from the MRI record collected
on Patient Case 01 from the Evaluation Dataset.
6 DISCUSSION
6.1 Approach for Pre-Processing
Qualitatively, the image after our contrast filter pre-
served distinctively more complete objects and shows
higher contrast. On the contrary, the denoised image
has a lower contrast after histogram equalization, and
the objects have missing parts.
Hence, Approach B, which applies our contrast
filter, is chosen and used for the pre-processing.
Reconstruction of 3D Brain Structures from Clinical 2D MRI Data
357
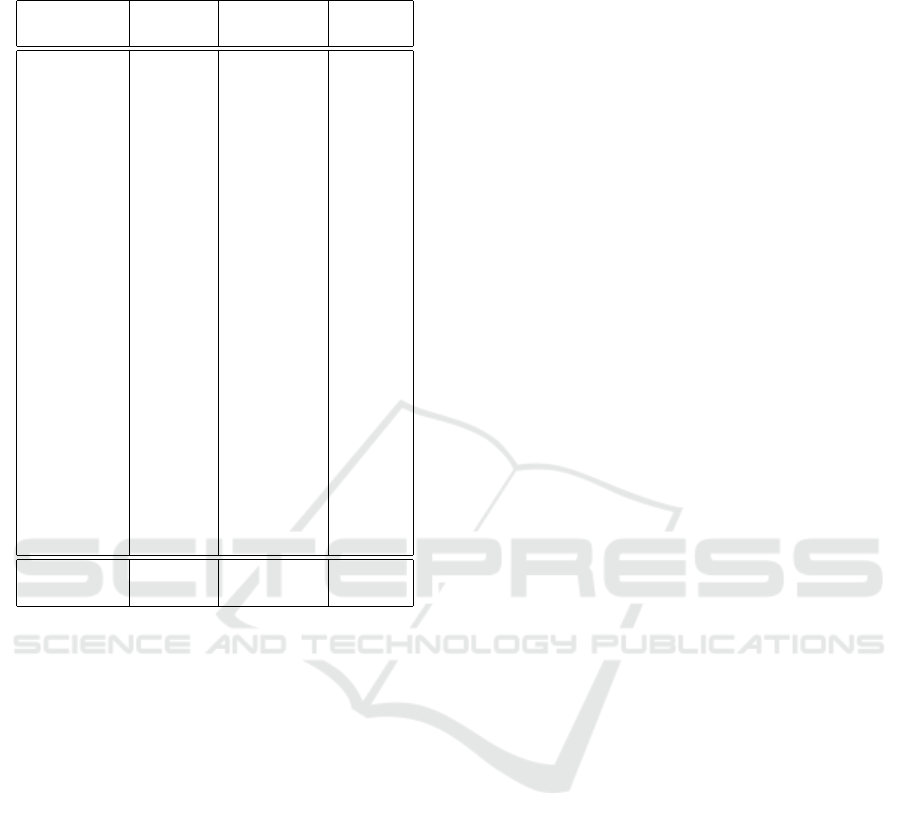
Table 1: Average PSNR and SSIM results for each patient
from the Evaluation Dataset.
Patient No. PSNR
SSIM SSIM
(Structure) (Slices)
01 22.3744 0.8900 0.5686
02 22.3910 0.8781 0.5633
03 21.3912 0.8632 0.5398
04 21.9015 0.8878 0.5623
06 23.3431 0.8752 0.5476
08 22.0405 0.8249 0.5362
09 22.3152 0.8981 0.5854
010 23.0736 0.8863 0.5773
012 22.6628 0.8423 0.5405
S1 22.8207 0.8808 0.5883
S2 22.5581 0.8906 0.5692
S3 22.2656 0.8381 0.5102
S4 24.0652 0.8618 0.5719
S5 23.0599 0.8792 0.5344
S6 20.7212 0.7676 0.5241
S7 19.3878 0.7326 0.4919
S8 22.5163 0.8869 0.5752
S9 24.1441 0.8653 0.5427
S13 22.7782 0.8797 0.5608
S14 22.4127 0.8727 0.5385
S15 22.3385 0.8202 0.5078
S16 23.3206 0.8918 0.5401
Mean 22.4492 0.8597 0.5489
Variance 1.0060 0.0017 0.0006
6.2 Adaptability, Stability and
Computational Speed
The proposed method shows high adaptability, suc-
ceeding in 3D brain structure generation based on all
three types of data scanned along different coordinate
axes in the Test Dataset, with images containing ap-
proximately 20 slices.
Performance stability can be perceived from 5b:
despite some deviations among the patient cases in
PSNR, only minor variance appears in SSIM (Struc-
ture) and SSIM (Slices) measurement.
The computation is also efficient. The average
time to generate the original 3D structure and the
reconstructed structure, and compute the PSNR and
SSIM values is within 2 seconds.
6.3 Pixel Similarity
The pixel similarity between the generated and the
original 3D structure is reflected by PSNR. The aver-
age PSNR value among all 22 cases of patients in the
Evaluation Dataset equals 22.45. Typical values for
the PSNR in lossy image and video compression are
between 30 and 50 dB, the higher the PSNR value, the
better quality the noisy image has (Faragallah et al.,
2020). Since there are fewer reference points in the
interpolation-generated images, the PSNR values are
generally lower (Jung and Yoo, 2009). Furthermore,
the objects inside the brain mainly consist of soft tis-
sues, which can be in small scales and various shapes,
yet have complicated spatial relationships (adjacent
and overlapping) (Nowinski, 2011). Considering the
delicacy of the brain structure, the obtained PSNR re-
sult and thus the pixel similarity are acceptable.
6.4 Structural Similarity
The structural similarity between the generated and
the original 3D structure is shown by SSIM (Struc-
ture). The average SSIM among the patients in the
Evaluation Dataset is 85.97%, with a minor vari-
ance of 0.2%. This indicates that the generated brain
structure can stably achieve 85.97% similarity to the
original 3D brain structure consisting of a relatively
large number of images (approx. 100), based on only
relatively few images (approx. 20, one-fifth of the
ground-truth). This achievement can be considered
inspiring.
The structural similarity between the generated
and the original 2D slices is reflected by SSIM (Slice).
The average SSIM among the patients is 54.89%,
along with a minor variance of 0.1%. This indicates
that the generated image slices can stably achieve
54.89% similarity to the original slices at the same
position. Unlike the remarkable resemblance in the
overview of the 3D structure, in a 2D slice perspec-
tive, the generated images lack much more similarity
to the original slices.
One possible explanation for the gap between the
2D and 3D performance could be that, although the
sampled data has large spacing intervals between the
slices, the slices still manage to cover the information
for the entire brain, among which the major charac-
teristics that matter in the 3D structure are still pre-
served. However, linear prediction cannot precisely
regenerate the details on a 2D slice within the empty
intervals, with limited information of that localized
area being only two adjacent slices.
7 CONCLUSION
We proposed a low-cost method to reconstruct the 3D
brain structure with routine clinical 2D MRI images,
using trilinear interpolation. The results indicate that
our method delivers good performance in 3D brain
structure reconstruction, achieving a similarity of up
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
358

to 85.97% between the original and the generated 3D
structure, using only one-fifth of the required amount
of images for the traditional stacking method.
As future work, to obtain better pixel resemblance
and more details in the generated 2D slices, further
improvement can be made with techniques such as
combinations of algorithms and assistance from neu-
ral networks. In the near future, our method may as-
sist doctors in making more precious diagnoses and
better treatments for diseases and injuries, even con-
quering dementia.
REFERENCES
Aja-Fern
´
andez, S. and Vegas-S
´
anchez-Ferrero, G. (2016).
Statistical analysis of noise in mri. Switzerland:
Springer International Publishing.
AlZu’bi, S., Shehab, M., Al-Ayyoub, M., Jararweh, Y., and
Gupta, B. (2020). Parallel implementation for 3d med-
ical volume fuzzy segmentation. Pattern Recognition
Letters, 130:312–318.
Bail, L. et al. (2009). The Human Head A Correct De-
lineation of the Anatomy, Expressions, Features, Pro-
portions and Positions of the Head and Face. Tom
Richardson.
Bathini, P., Brai, E., and Auber, L. A. (2019). Olfactory
dysfunction in the pathophysiological continuum of
dementia. Ageing research reviews, 55:100956.
Bull, D. R. and Zhang, F. (2014). Digital picture formats
and representations. Communicating pictures, pages
99–132.
Carlson, N. R. and Carlson, N. R. (2007). Physiology of
behavior. Pearson Boston.
Davis, P. J. (1975). Interpolation and approximation.
Courier Corporation.
Ebel, K.-D. and Benz-Bohm, G. (1999). Differential diag-
nosis in pediatric radiology. Thieme.
Fajar, A., Sarno, R., Fatichah, C., and Fahmi, A. (2022). Re-
constructing and resizing 3d images from dicom files.
Journal of King Saud University-Computer and Infor-
mation Sciences, 34(6):3517–3526.
Faragallah, O. S., El-Hoseny, H., El-Shafai, W., Abd El-
Rahman, W., El-Sayed, H. S., El-Rabaie, E.-S. M.,
Abd El-Samie, F. E., and Geweid, G. G. (2020). A
comprehensive survey analysis for present solutions
of medical image fusion and future directions. IEEE
Access, 9:11358–11371.
Ghoshal, S., Banu, S., Chakrabarti, A., Sur-Kolay, S., and
Pandit, A. (2020). 3d reconstruction of spine image
from 2d mri slices along one axis. IET Image Pro-
cessing, 14(12):2746–2755.
Jung, K.-H. and Yoo, K.-Y. (2009). Data hiding method
using image interpolation. Computer Standards & In-
terfaces, 31(2):465–470.
Juntu, J., Sijbers, J., Van Dyck, D., and Gielen, J. (2005).
Bias field correction for mri images. In Computer
Recognition Systems: Proceedings of the 4th Interna-
tional Conference on Computer Recognition Systems
CORES’05, pages 543–551. Springer.
Lorensen, W. E. and Cline, H. E. (1998). Marching cubes:
A high resolution 3d surface construction algorithm.
In Seminal graphics: pioneering efforts that shaped
the field, pages 347–353.
Mustra, M., Delac, K., and Grgic, M. (2008). Overview of
the dicom standard. In 2008 50th International Sym-
posium ELMAR, volume 1, pages 39–44. IEEE.
Nowinski, W. L. (2011). Introduction to brain anatomy.
Biomechanics of the Brain, pages 5–40.
Patel, A. and Mehta, K. (2012). 3d modeling and rendering
of 2d medical image. In 2012 International Confer-
ence on Communication Systems and Network Tech-
nologies, pages 149–152. IEEE.
Rajon, D. A. and Bolch, W. E. (2003). Marching cube algo-
rithm: review and trilinear interpolation adaptation for
image-based dosimetric models. Computerized Medi-
cal Imaging and Graphics, 27(5):411–435.
Salerno, J. A., Murphy, D., Horwitz, B., DeCarli, C.,
Haxby, J. V., Rapoport, S. I., and Schapiro, M. B.
(1992). Brain atrophy in hypertension. a volumet-
ric magnetic resonance imaging study. Hypertension,
20(3):340–348.
Senthilkumaran, N. and Thimmiaraja, J. (2014). Histogram
equalization for image enhancement using mri brain
images. In 2014 World congress on computing and
communication technologies, pages 80–83. IEEE.
Statistics Bureau (2023). Population and households. In
JAPAN STATISTICAL YEARBOOK 2024, chapter 2,
pages 29–82. Ministry of Internal Affairs and Com-
munications, Tokyo.
Thanh, C. Q. and Hai, N. T. (2017). Trilinear interpolation
algorithm for reconstruction of 3d mri brain image.
American Journal of Signal Processing, 7(1):1–11.
van der Flier, W. M. and Scheltens, P. (2005). Epidemiology
and risk factors of dementia. Journal of Neurology,
Neurosurgery & Psychiatry, 76(suppl 5):v2–v7.
Wang, Z., Bovik, A. C., Sheikh, H. R., and Simoncelli, E. P.
(2004). Image quality assessment: from error visi-
bility to structural similarity. IEEE transactions on
image processing, 13(4):600–612.
Reconstruction of 3D Brain Structures from Clinical 2D MRI Data
359
