
Pilot Study of Distinct Graphs Models in Analysis of Brain Aging in
Resting-State Functional Connectivity Networks
M. A. G. Carvalho
1,2,3 a
and R. Frayne
2,3,4,5 b
1
School of Technology, University of Campinas, Brazil
2
Radiology and Clinical Neurosciences, Hotchkiss Brain Institute, Univ. of Calgary, Canada
3
Seaman Family MR Research Centre, Foothills Medical Centre, Calgary, Canada
4
Biomedical Engineering Graduate Program, University of Calgary, Canada
5
Calgary Image Processing and Analysis Centre, Foothills Medical Centre, Calgary, Canada
Keywords:
Graph Model (GM), Graph Metrics, Resting-State Functional Magnetic Resonance Imaging, Brain Networks,
Aging.
Abstract:
Graphs have been used successfully to represent and analyze brain networks for many decades. Such structural
and functional studies are important for revealing interactions between distinct areas of the brain that, for ex-
ample, are associated with the performance of a specific task or the onset of a cognitive disorder like dementia.
In this pilot study, resting-state functional magnetic resonance imaging data were acquired in a sex-balanced
sample of 10 young (20.1 ± 2.1 years) and 10 old (65.6 ± 0.4 years), presumed healthy, adults. We examined
the effects of age on whole-brain resting-state functional connectivity (RSFC) networks. We examined two
main graph modeling approaches to analyze RSFC networks. These approaches employ different strategies or
graph models for thresholding over the complete network or examining changes in graph density. We com-
puted and compared one graph metric, the modularity, that was derived from the RSFC network graph models.
Considering the need for a model that must preserve the network’s connectivity, strategies that use spanning
trees as seeds to gradually increase the graph’s density seem more appropriate to represent brain networks.
1 INTRODUCTION
Neuroimaging techniques have become more accessi-
ble and frequently used in recent years and have facil-
itate the emergence of human brain studies, including
investigations of brain aging. Such aging studies at-
tempt to understand how the healthy brain transforms
morphologically or functionally across the lifespan.
Considering that in many countries people are living
longer, it is important to first study healthy aging if
we are to then attempt to understand pathological ag-
ing. This knowledge can then help understand and
mitigate impacts of illnesses like cognitive loss and
dementia. Functional magnetic resonance imaging
(fMRI) is a widely employed, non-invasive method
to investigate functional organization of and organi-
zational change in the brain during aging. Resting-
state fMRI (rs-fMRI) methods process the sponta-
neous fluctuations in the blood oxygen levels (or the
a
https://orcid.org/0000-0002-1941-6036
b
https://orcid.org/0000-0003-0358-1210
BOLD signal - blood oxygen level–dependent) that
occur even in the absence of a stimulus or activation
task (Lv et al., 2018).
To describe the brain functional organization, i.e.,
its underlying functional connectivity, studies have
modeled the brain as a highly structured network,
known as the connectome (Hrybouski et al., 2021).
Graph theory approaches naturally arise as a method
for representing human brain networks. In the mathe-
matical sense, a graph G is an ordered pair G = (V,E)
defined by a set of nodes (or vertices) V and set
of edges (or links) E connecting the nodes (Chung,
2019). The use of graphs to represent networks is im-
portant as it enables characterization and analysis of
brain functional organization using a variety of graph
metrics.
This initial study aims to explore the computation
of modularity graph metric according to distinct graph
models (GMs). Such metric characterize relevant as-
pect of resting-state functional connectivity (RSFC)
networks. We conducted an experiment using four
348
Carvalho, M. A. G. and Frayne, R.
Pilot Study of Distinct Graphs Models in Analysis of Brain Aging in Resting-State Functional Connectivity Networks.
DOI: 10.5220/0013261200003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 348-356
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
GMs, corresponding to representations for RFSC net-
works, widely reported in the literature. This work
makes contributions in the following areas:
• We present an up-to-date review of the literature
of GM representations of the RSFC networks.
• We analyzed RSFC networks using a graph metric
calculated for different GMs in order to improve
understanding of the potential and limitations of
each model.
The remainder of this paper is organized as fol-
lows: In Section 2 the literature survey protocol is
presented. We then summarize our findings regard-
ing the graph representation used to model RSFC net-
works. Section 3 describes our experiment method-
ology, expanding on the important details required to
build distinct GMs. Section 4 summarizes our results
and discusses the broader implications of our find-
ings. Finally, Section 5 concludes the paper with final
remarks and a discussion of remaining issues.
2 LITERATURE REVIEW
In this section we explain how we identify key works
in the peer-reviewed literature on RSFC brain net-
works of healthy individuals.
We used the three-staged methodology in order to
select relevant papers that explored graph theory, i.e.,
uses GMs to characterize and analyze RSFC networks
in aging (Figure 1).
Figure 1: Three-stage protocol to select papers dealing with
the representation of RSFC networks through graph models.
The paper extraction intended to answer the fol-
lowing questions:
1. What are the main graph approaches used to rep-
resent RSFC networks?
2. Concerning the identified approaches, what are
the strategies or variations regarding to the graph
models?
3. What other graph properties (or graph types) are
important in characterizing RSFC networks?
2.1 Exploring Existing Reviews
In this stage we collect three recent review or system-
atic review papers, listed in Table 1, that works with
graph theory to characterize and analyze RSFC net-
works in aging.
The three review and systematic review papers
identified a number of broad-based studies focused on
brain aging in healthy individuals that analyzed RSFC
networks. Combined, they cited a total of twenty-
six papers that specified modeling of RSFC networks
using graph theory. Only paper that explicitly men-
tioned using graph metrics were included. We ex-
cluded reports that focused on addressing specific dis-
eases, trauma or that studied the effects of medica-
tions, other treatments or therapies. We also excluded
reports not written in English. Twenty-five of these
papers were selected (see list in Table 2) and one arti-
cle was excluded (one study obtained rs-fMRI data in
hypercapnia (Hou et al., 2019)).
2.2 Supplementary Screening and
Inclusion/Exclusion Criteria
Because the most recent of the three review and
systematic review examined papers published before
June 2021 (Deery et al., 2023), a supplementary
search was conducted, according to a three-step pro-
tocol as described in (Kitchenham, 2004), in order to
include articles between June 2021 and June 2024.
The supplementary search was performed using
two complementary databases: 1) IEEE Xplore
1
(en-
gineering and computer science literature) and 2)
PubMed
2
(biomedicine literature) using the research
query: “(aging) AND (brain) AND (network) AND
(functional connectivity) AND (review OR systematic
review)”. The inclusion criteria initially considered
only papers focusing on brain aging evaluated with
RSFC networks analysis.
Were used the same inclusion and exclusion crite-
ria applied in previous stage. Finally, we have iden-
tified an additional six papers published after May
2021.
2.3 Studies: Identification and
Summary
These 31 selected articles, including both stages,
cover a period between 2007 and 2024, and constitute
our source of primary studies. Those papers were an-
alyzed in detail, specifically examining the GM used.
After analysis, we have classified each approach
concerning the use of graph theory on three types
of graph (GT - Graph Type) and four types of
graph model (GM) (see Table 2). The GTs were:
1
https://ieeexplore.ieee.org/Xplore/home.jsp
2
https://pubmed.ncbi.nlm.nih.gov/
Pilot Study of Distinct Graphs Models in Analysis of Brain Aging in Resting-State Functional Connectivity Networks
349
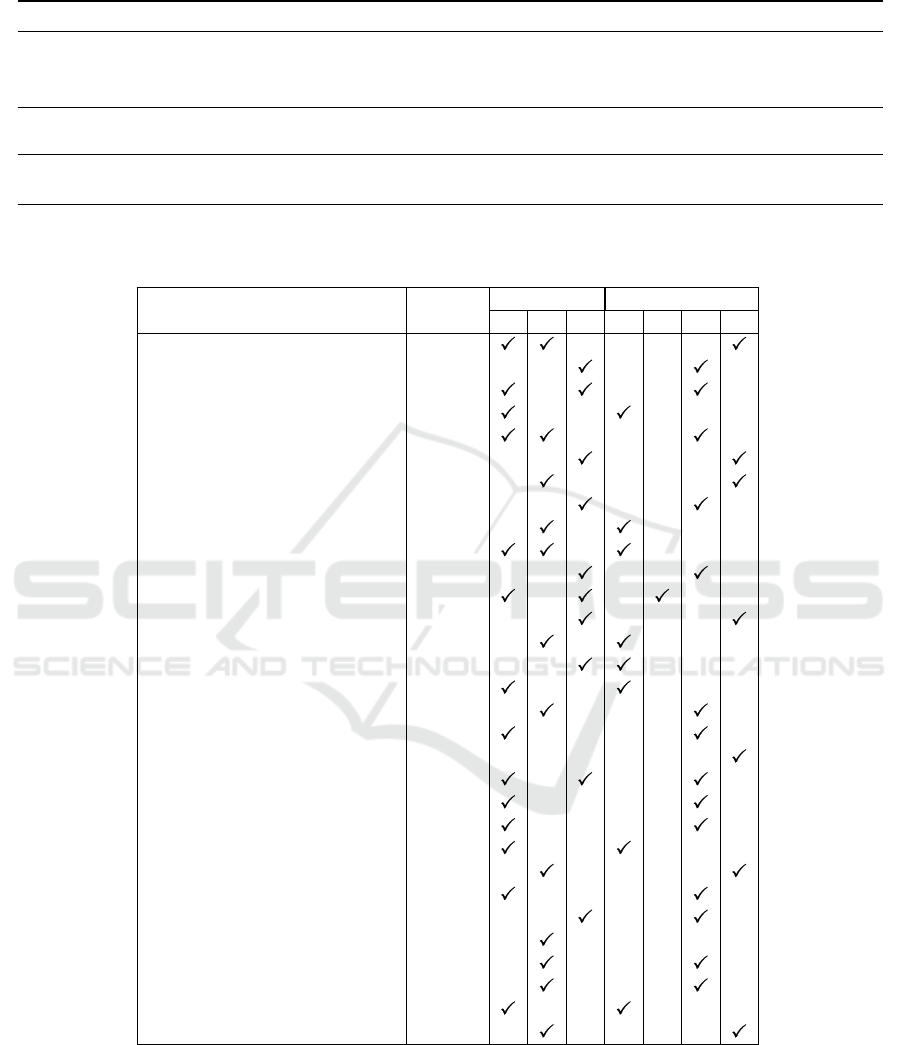
Table 1: Review and systematic review papers selected on stage 1.
Year Paper Title Ref
2023 The older adult brain is less modular, more integrated, and less
efficient at rest: A systematic review of large-scale resting-state
functional brain networks in aging
(Deery et al., 2023)
2021 Resting-state networks in the course of aging-differential in-
sights from studies across the lifespan vs. amongst the old
(Jockwitz and
Caspers, 2021)
2020 Functional brain connectivity changes across the human life
span: From fetal development to old age
(Edde et al., 2020)
Table 2: Summary of graph based methods, summarizing study design, graph type (GT) and graph model (GM). Publications
extracted from Ref (Deery et al., 2023),(Jockwitz and Caspers, 2021), and (Edde et al., 2020), and updated to June 2024.
Study GT GM
Ref Design 1 2 3 1 2 3 4
(Hrybouski et al., 2021) CS - - - -
(Grady et al., 2016) CS - - - - -
(Geerligs et al., 2015) CS - - - -
(Gallen et al., 2016) CS - - - - -
(Iordan et al., 2018) CS - - - -
(Betzel et al., 2014) CS - - - - -
(Chan et al., 2014) CS - - - - -
(Stumme et al., 2020) CS - - - - -
(Alcauter et al., 2015) CS - - - - -
(Asis-Cruz et al., 2015) CS - - - -
(Marek et al., 2015) CS - - - - -
(Song et al., 2014) CS - - - -
(Thomason et al., 2014) CS - - - - -
(van den Heuvel et al., 2018) CS - - - - -
(Varangis et al., 2019) CS - - - - -
(Bagarinao et al., 2019) CS - - - - -
(Cao et al., 2014) CS - - - - -
(Li et al., 2016) CS - - - - -
(Onoda and Yamaguchi, 2013) CS - - - - - -
(Sala-Llonch et al., 2014) CS - - - -
(Meunier et al., 2009) CS - - - - -
(Achard and Bullmore, 2007) CS - - - - -
(Lehmann et al., 2021)
∗
CS - - - - -
(Foo et al., 2021)
∗
CS - - - - -
(Wang et al., 2024)
∗
CS - - - - -
(Moretto et al., 2022)
∗
CS - - - - -
(Filippi et al., 2023)
∗
CS - - - - -
(Chong et al., 2019) CS/L - - - - -
(Wen et al., 2019) L - - - - -
(Xiao et al., 2016) L - - - - -
(Pedersen et al., 2021)
∗
L - - - - -
∗
= Additional paper identified in supplementary screening
GTs were: GT1-binary graph, GT2-weighted graph, only positive
values, and GT3-weighted graph, positive and negative values.
GMs were: GM1-thresholding operation approach, GM2-spanning tree
initial seed approach, GM3-density approach, and GM4-entire graph.
CS = Cross-Sectional; L = Longitudinal
BIOIMAGING 2025 - 12th International Conference on Bioimaging
350

GT1) binary graph (14/31, 45.2%), GT2) weighted
graph, only positive values (12/31, 38.7%), and GT3)
weighted graph, positive and negative values (10/31,
32.2%). Some papers work with multiple GT, which
can result in a sum greater than 30. The use of bi-
nary graphs was associated with the evaluation of as-
pects of the network topology. This observation is
related to the scope of the study, i.e., being able to
evaluate only aspects of the network topology, as in
the case of using binary graphs, or when it was neces-
sary to evaluate more specific graph properties, such
as strength and shortest path length. The detailed re-
view also identified an alternative GM aimed at inves-
tigating brain hubs, i.e., highly connected regions of
the neurocognitive functional networks (Filippi et al.,
2023). This approach was recently used to obtain new
adjacency matrices from the analysis of the distance
of any brain region, according to the degree of step-
wise connectivity and the seed area (i.e., hubs).
The models included GM1) based on a threshold-
ing operation (fixed value or range) (8/31, 25.8%),
GM2) based on the construction of a spanning tree
and the addition of successive edges (1/31, 3.2%),
GM3) based on the pre-defined values of graph den-
sity range (14/31, 45.2%), and GM4) based on the
entire and non-sparse graph (7/31, 22.6%).
The first model (GM1) used thresholding. One of
the main challenges in applying strategies that use a
thresholding process is defining the threshold itself, as
this operation may include irrelevant or disregard rel-
evant information from the RSFC network (van den
Heuvel et al., 2017). One way to mitigate the im-
pact of using a single threshold is to apply a range
of values. Indeed, most studies use a threshold range
(Asis-Cruz et al., 2015)(van den Heuvel et al., 2018)
(Chong et al., 2019). In Bagarinao (Bagarinao et al.,
2019), for example, a threshold range [0.20,0.40] was
used. We also found studies that used a single thresh-
old (Alcauter et al., 2015)(Xiao et al., 2016). One
aspect that is not always explicitly reported was the
fact that the thresholding process can generate dis-
connected RSFC networks. In this situation, the cal-
culation of graph metrics were performed separately
on either each connected component or only on the
largest connected component.
GM2 derives a spanning tree (MST - minimum
spanning tree in that case) as a seed and gradually
include a percentage of edges, as adopted by (Song
et al., 2014). This method represents an interest-
ing approach as it guarantees the connectivity of the
graph. One important point is that the weights of the
edges added in a minimum spanning tree are those
with smaller values, in ascending order, according to
Kruskal’s algorithm (Fornito et al., 2016). Consider-
ing the graph density approach added relevant edges,
i.e., stronger connections, in this case, therefore, it
would be appropriate to work with a maximum span-
ning tree (MaxST).
GM3 consists of defining a range of graph den-
sity values (Grady et al., 2016) (Marek et al., 2015)
and generating representations of the RSFC network.
Graph density is a measure of the ratio between edges
and nodes. The density is 0 for a graph without edges
and 1 for a complete graph. The graph density strat-
egy is similar to spanning trees because with the ex-
ception of not using a seed, it also consists of gradu-
ally adding relevant edges. One limitation is the pos-
sibility of creating disconnected graphs. We found
papers that worked with graph densities that ranged
between 1% and 60% (Grady et al., 2016) and 1%
and 25% (Marek et al., 2015).
The final model (GM4) used the entire graph, such
that each entry of the Pearson correlation matrix cor-
responds to an edge in the graph. In (Thomason et al.,
2014), for instance, both positive and negative corre-
lation values were used in order to build the equiva-
lent graph. On the other hand, (Chan et al., 2014) only
positive values of the correlation matrix were used.
Table 3 list the five most cited articles listed in
Table 2. The most cited of these papers presented
the calculation of local and global efficiency graph
metrics, in addition to using a graph model approach
based on graph density (Achard and Bullmore, 2007).
Table 3: Highly cited papers (Source: PubMed).
Ref Total Citations
citations per year
(Achard and Bullmore, 2007) 1113 > 60
(Chan et al., 2014) 399 ≈ 40
(Betzel et al., 2014) 392 ≈ 40
(Geerligs et al., 2015) 348 ≈ 40
(Meunier et al., 2009) 370 ≈ 25
2.4 Considerations on Review Findings
The connectivity of a GM is an important feature in
the analysis of RSFC networks. Graph metric compu-
tation may undergo changes according to the connec-
tivity property, such as corresponding to the value of
the largest connected component. Iordan et al.(Iordan
et al., 2018), for instance, explicitly pointed out the
number of nodes that were disconnected in the graphs
built in his modeling. Meunier et al.(Meunier et al.,
2009) defined the number of connections, in a range
between 100 and 400 edges (or links), in order to
keep the graph connected. An interesting strategy
that guarantees the connectivity of a graph model
is the use of minimum spanning trees (Song et al.,
Pilot Study of Distinct Graphs Models in Analysis of Brain Aging in Resting-State Functional Connectivity Networks
351

2014). A spanning tree is a tree-like subgraph of
a connected graph that includes all vertices. More
consistent results may be found when employing a
maximum spanning tree, instead minimum spanning
tree, because the most relevant edges of the graph are
added in sequence.
To correctly understand the brain aging process
trajectory, it is necessary to explore longitudinal
data. This methodology investigates changes in intra-
individual functional connectivity networks and al-
lows us to understand the compensatory changes car-
ried out by the different RSFC networks in healthy in-
dividuals. Only a few studies (4/30, 13.3%) used lon-
gitudinal data when calculating graph metrics (Chong
et al., 2019)(Wen et al., 2019)(Xiao et al., 2016)(Ped-
ersen et al., 2021). These four studies only exam-
ined one age group and, therefore, have limitations
in terms of extrapolation of results over the adult life
span.
Choosing a GM is not just a question of efficiency,
as they all enable RSFC network characterization us-
ing graph metrics via appropriate parameter defini-
tion. Choosing a GM is about knowing the condi-
tions for acquiring and processing rs-fMRI data and
the study sample, avoiding spurious connections and
biases (Rubinov and Sporns, 2010)(Varangis et al.,
2019).
We also observed the existence of distinct nomen-
clatures to represent the number or percentage of
edges in the graph model, as well as in the definition
of relevant entries (correlation values) of the FC ma-
trix. Considering that the origin of the term thresh-
olding is related to the modification of intensity or
amplitude values in a matrix, we propose to use the
simple terminology for the GMs found in the litera-
ture. The described GMs and graph analysis scenarios
can be organized into two major approaches for ana-
lyzing RSFC networks: 1) Thresholding, where the
weighted edges of the corresponding graph must be
greater than a value or a range of values, and 2) Graph
density, where the resulting graph is constructed from
a percentage of connections in the original network,
including the entire graph.
3 METHODS
The modularity analysis pipeline is shown in Fig. 2
and consisted of four main steps described in this sec-
tion.
Figure 2: Overview of processing pipeline.
3.1 Data Acquisition
MR imaging data in this study were acquired as part
of the Calgary Normative Study (CNS) (McCreary
et al., 2020). The CNS is an ongoing longitudi-
nal study, started in 2013, that focuses on collect-
ing quantitative data from community dwelling, pre-
sumed healthy adults (18-90+ years). Several types
of MR neuroimaging were performed in the CNS in-
cluding rs-fMRI. To pilot our methods, we exam-
ined twenty (20) individuals extracted from the CNS
database, ten (10) from a young and ten (10) from an
older group. Table 4 lists the demographics of the
groups.
Table 4: Dataset sample demographics.
Group (Number) Sex Ratio Age (years)
Young (N = 10) 50% Female 20.1 ± 2.1
50% Male
Old (N = 10) 50% Female 65.6 ± 0.4
50% Male
3.2 Image Preprocessing
The preprocessing pipeline consists of the prepara-
tion and analysis of the rs-fMRI images. The pipeline
comprises several key steps: 1) skull striping us-
ing BET, 2) motion correction using MCFLIRT (Mo-
tion Correction FMRIB Linear Image Registration
Tool), 3) interleaved slice-time correction, 4) spatial
smoothing, 5) temporal high-pass filtering, 6) inde-
pendent component analysis (ICA) and 7) functional-
structural registration. Details of the pipeline are de-
scribed in (Sidhu, 2023).
3.3 FC Network Matrices and Graph
Models
From the average BOLD time series, a FC matrix of
size 200×200 was derived for each individual by cal-
culating the Pearson correlation (r) across the time
series (Sidhu, 2023). This matrix size was obtained
BIOIMAGING 2025 - 12th International Conference on Bioimaging
352

from the brain segmentation into 200 anatomical re-
gions using the Schaefer-Yeo atlas. We consider a
brain organization based on the existence of seven rel-
evant modules (Sidhu, 2023): Visual network, Senso-
rimotor network, Frontoparietal network, Dorsal at-
tention network, Limbic network, Ventral attention
network, Default mode network. After analyzing the
quality of the rs-fMRI data, FC matrix entries from
the left and right limbic networks were excluded be-
cause of signal loss resulting from MR susceptibility
artifacts. Fisher’s r–to–z transformation was then ap-
plied to the Pearson correlation values and the main
diagonal of the FC matrix was set to zero to exclude
self connections.
We explored four GMs that can be used to repre-
sent RSFC networks from FC matrices. These GM
were first identified in our systematic review (Table
2):
• GM 1: Thresholding applied a threshold range
directly to the Pearson correlation matrix.
• GM 2: Minimum Spanning Tree was built from
all graphs. Graph connectivity was preserved
and edges were added according to their weight,
choosing first to add those with the lower values.
• GM 3: Graph Density used a range of values
for the graph density metric. We gradually add
edges (as a percentage) to the model graph, as in
the previous model.
• GM4: Entire Graph used all positive values of
the Pearson correlation matrix. Each matrix entry
corresponds to an edge of the graph.
We decided to include a fifth GM, GM5, corre-
sponding to a variation of spanning tree called Maxi-
mum Spanning Tree. Unlike the Minimum spanning
tree, this tree is built by considering the strongest con-
nections, as done by the thresholding and graph den-
sity approaches. In that case, edges are added accord-
ing to their weight, choosing first to add those with the
higher values. In all cases where a range was used, the
final value of the graph metric was the average value
calculated over that range.
3.4 Modularity: Graph Metric
Modularity (Q) measures the relative strength of a
network division into groups. Modularity reflects the
existence of subnetworks within the full network.
Q =
∑
I∈M
l
II
−
∑
J∈M
l
IJ
!
2
(1)
where the network is divided into a set of nonoverlap-
ping modules M and l
IJ
is the ratio of all links that
connect nodes in module I with module J. RSFC net-
works with high modularity have dense connections
between nodes within the module but sparse connec-
tions between nodes across different modules. Details
concerning modularity can be found at (Fornito et al.,
2016).
4 RESULTS AND DISCUSSIONS
Our objective was to highlight trends in the five GMs
between the young and old age groups and com-
pare them with literature findings. For all exper-
iments, we use Python and the networkX library
(https://networkx.org/).
The GMs examined were (section 3.3): GM1 –
thresholding with fifteen steps in the range [0.05,0.4];
GM2 and GM5 – minimum and maximum spanning
trees followed by a gradual addition of edges in the
range of 2% and 40% (Song et al., 2014); GM3 –
graph density using the range [1%,25%]; and GM4
– entire graph(Marek et al., 2015). For all models,
we processed only positive Pearson coefficient values
and computed the modularity (section 3.4) to evalu-
ate age-related change in the GMs. Figure 3 presents
results for all examined GMs.
As we can see in Figure 3, Q decreased in the
old compared to the young group, over a range of dif-
ferent thresholds and edges densities. This was also
observed in GM4, where the mean values obtained for
the young and old groups were 0.1162 and 0.1099, re-
spectively. This finding is similar to that obtained in
Deery (Deery et al., 2023) who found that in 100% of
studies Q decreased with age.
In the neuroimaging field, the construction of
networks and their analyses draws on the concept of
small world networks (Deery et al., 2023). Small-
world networks are defined as networks that are sig-
nificantly more clustered than random networks (Ru-
binov and Sporns, 2010). Except for the GM4 model,
all GMs presents mean modularity values greater than
0.3, an indicative of nonrandom community structure,
i.e., the existence of modular structure of functional
brain networks across the adult lifespan(Song et al.,
2014). Finally, Minimum and Maximum spanning
trees (GM2 and GM5, respectively) present a simi-
lar behavior; however, considering that GM5 includes
first the most relevant connections in terms of Pearson
correlation values, this would be the most coherent
choice.
Pilot Study of Distinct Graphs Models in Analysis of Brain Aging in Resting-State Functional Connectivity Networks
353
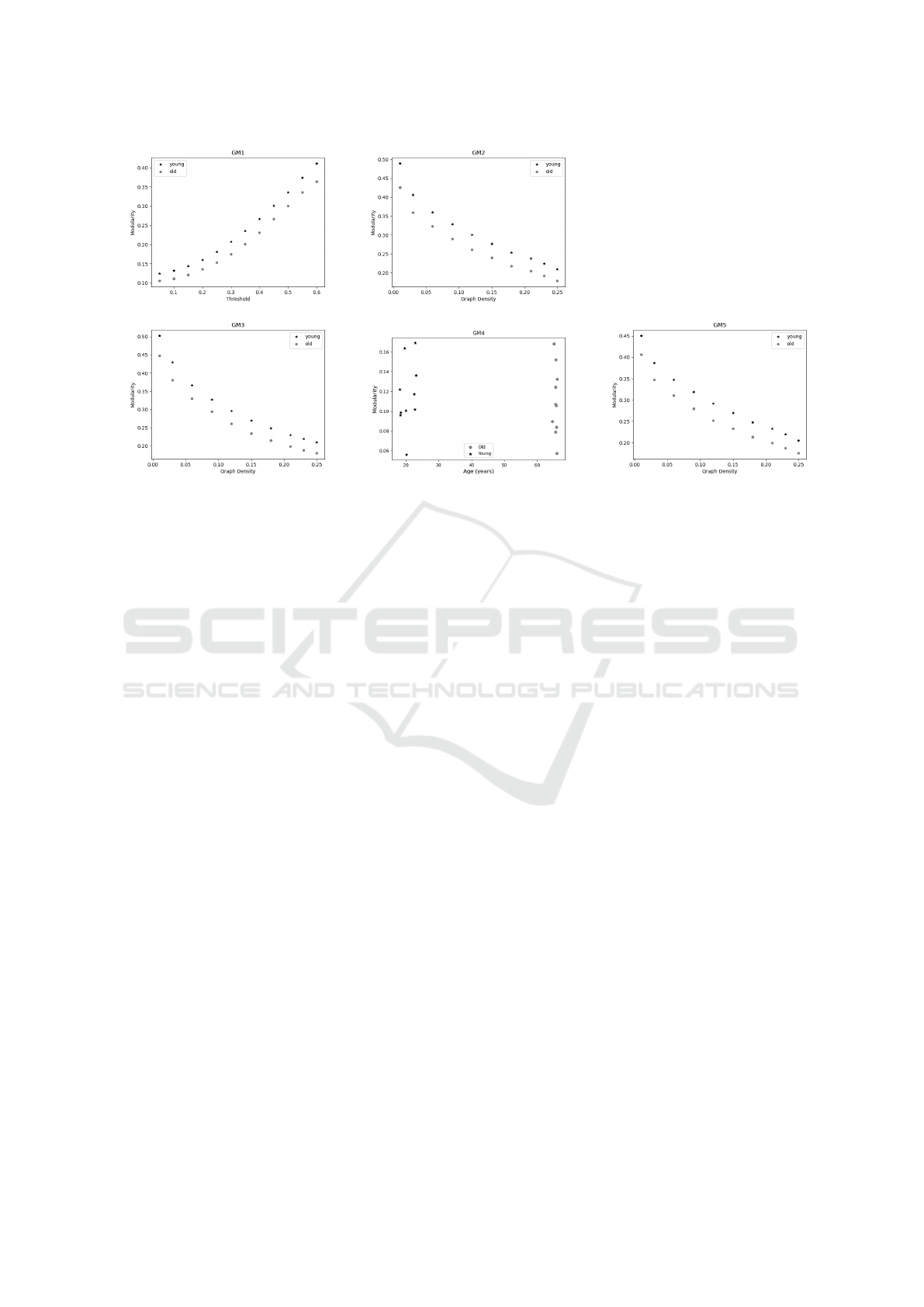
Figure 3: Modularity values by graph model. for young (black stars) and old individuals (gray circles). Rows are graph
models: GM1, GM2, and GM3, GM4 and GM5 (top to bottom).
5 CONCLUSIONS
We found thirty peer-reviewed papers that addressed
changes in RSFC networks in aging using graphs (Ta-
ble 2. Many additional papers used graphs to study
diseases and disorders, but were outside our selection
criteria. It was possible to identify two approaches
to model brain as graphs: Thresholding operations
(that act on the Pearson correlation values), and ap-
proaches that use a gradual increase in graph density
(that act on the number of edges or links). More stud-
ies only worked with positive Pearson correlation val-
ues, than those using positive and negative values. In
addition, binary graphs were widely used to express
network characteristics, such as modularity and clus-
tering coefficient. Also of note is the maintenance of
the graph connectivity property, observed from its ad-
jacency matrix. Graph connectivity is important be-
cause it allows the application of other mathematical
approaches and more advanced computational tech-
niques. In this context, the model that uses spanning
trees naturally results in connected graphs.
We carried out a pilot experiment that com-
pared a common graph metric (modularity), com-
puted over distinct graph models representing RSFC
networks, to analize existing differences among
groups of young and old healthy individuals. This ex-
periment presents findings similar to those obtained
in the literature, which indicate that the value of mod-
ularity decreases with aging. We highlight that this
work consists of an exploratory analysis of GMs. An
in-depth study of the full CNS dataset with respect to
brain aging including other parameters is warranted.
Finally, to understand the brain aging process from
a different perspective, we should examine longitu-
dinal data, that is, study changes in intra-individual
functional connectivity networks, considering age as
a continuous variable. This type of study will also al-
low us to understand changes made in different RSFC
networks over age in healthy individuals.
ACKNOWLEDGEMENTS
This study was financed in part by the Coordination of
Improvement of Higher Education Personnel - Brazil
(CAPES) - Finance Code 001. Marco Carvalho wish
to express their gratitude to the S
˜
ao Paulo Research
Foundation/Fundac¸
˜
ao de Amparo
`
a Pesquisa do Es-
tado de S
˜
ao Paulo (FAPESP grant 2023/02302-6). We
also acknowledge the assistance of Kau
ˆ
e TN Duarte,
Abhi S Sidhu, and Cherly R McCreary, from Univer-
sity of Calgary.
REFERENCES
Achard, S. and Bullmore, E. (2007). Efficiency and cost of
economical brain functional networks. PLoS Compu-
tational Biology, 3(2):e17.
Alcauter, S., Lin, W., Keith Smith, J., Gilmore, J. H., and
Gao, W. (2015). Consistent anterior-posterior segrega-
BIOIMAGING 2025 - 12th International Conference on Bioimaging
354

tion of the insula during the first 2 years of life. Cere-
bral Cortex, 25:1176–1187.
Asis-Cruz, J. D., Bouyssi-Kobar, M., Evangelou, I., Vezina,
G., and Limperopoulos, C. (2015). Functional proper-
ties of resting state networks in healthy full-term new-
borns. Scientific Reports, 5:17755.
Bagarinao, E., Watanabe, H., Maesawa, S., Mori, D.,
Hara, K., Kawabata, K., Yoneyama, N., Ohdake, R.,
Imai, K., Masuda, M., Yokoi, T., Ogura, A., Taoka,
T., Koyama, S., Tanabe, H. C., Katsuno, M., Wak-
abayashi, T., Kuzuya, M., Ozaki, N., and Sobue, G.
(2019). Reorganization of brain networks and its as-
sociation with general cognitive performance over the
adult lifespan. Scientific Reports, 9(1):11352.
Betzel, R. F., Byrge, L., He, Y., Goni, J., Zuo, X. N.,
and Sporns, O. (2014). Changes in structural and
functional connectivity among restingstate networks
across the human lifespan. Neuroimage, 102:345–
357.
Cao, M., Wang, J. H., Dai, Z. J., Cao, X. Y., Jiang, L. L.,
Fan, F. M., Song, X. W., Xia, M. R., Shu, N., Dong,
Q., Milham, M. P., Castellanos, F. X., Zuo, X. N., and
He, Y. (2014). Topological organization of the human
brain functional connectome across the lifespan. De-
velopmental Cognitive Neuroscience, 7:76–93.
Chan, M. Y., Park, D., Savalia, N., Petersen, S., and Wig, G.
(2014). Decreased segregation of brain systems across
the healthy adult lifespan. Proc Natl Acad Sci U S A,
111:E4997–E5006.
Chong, J., Ng, K. K., Tandi, J., Wang, C., Poh, J. H., Lo,
J. C., Chee, M., and Zhou, J. H. (2019). Longitudi-
nal changes in the cerebral cortex functional organiza-
tion of healthy elderly. The Journal of Neuroscience,
39(28):5534–5550.
Chung, M. K. (2019). Brain Network Analysis. Cambridge
University Press.
Deery, H. A., Di Paolo, R., Moran, C., , Egan, G. F., and
Jamadar, S. D. (2023). The older adult brain is less
modular, more integrated, and less efficient at rest:
A systematic review of large-scale resting-state func-
tional brain networks in aging. Psychophysiology,
60:e14159.
Edde, M., Leroux, G., Altena, E., and Sandra, C. (2020).
Functional brain connectivity changes across the hu-
man life span: From fetal development to old age.
Journal of Neuroscience Research, 99(1):236–262.
Filippi, M., Cividini, C., Basaia, S., Spinelli, E. G., Castel-
novo, V., Leocadi, M., Canu, E., Agosta, F., and
Bertoldo, A. (2023). Age-related vulnerability of
the human brain connectome. Molecular Psychiatry,
28:5350–5358.
Foo, H., Thalamuthu, A., Jiang, J., Koch, F., Mather, K. A.,
Wen, W., and Sachdev, P. S. (2021). Age- and sex-
related topological organization of human brain func-
tional networks and their relationship to cognition.
Frontiers in Aging Neuroscience, 13:758817.
Fornito, A., Zaleski, A., and Bullmore, E. T. (2016). Fun-
damentals of brain network analysis. Elsevier.
Gallen, C. L., Turner, G. R., Adnan, A., and D’Esposito, M.
(2016). Reconfiguration of brain network architecture
to support executive control in aging. Neurobiology of
Aging, 44:42–52.
Geerligs, L., Renken, R., Saliasi, E., Maurits, N., and Lorist,
M. (2015). A brain wide study of age-related changes
in functional connectivity. Cereb Cortex, 25:1987–
1999.
Grady, C., Sarraf, S., Saverino, C., and Campbell, K.
(2016). Age differences in the functional interactions
among the default, frontoparietal control, and dorsal
attention networks. Neurobiol Aging, 41:159–172.
Hou, X., Liu, P., Gu, H., Chan, M., Li, Y., Peng, S. L., Wig,
G., Yang, Y., Park, D., and Lu, H. (2019). Estima-
tion of brain functional connectivity from hypercapnia
bold mri data: Validation in a lifespan cohort of 170
subjects. Neuroimage, 186:455–463.
Hrybouski, S., Cribben, I., McGonigle, J., Olsen, F., Carter,
R., Seres, P., Madan, C. R., and Malykhin, N. V.
(2021). Investigating the effects of healthy cogni-
tive aging on brain functional connectivity using 4.7 t
resting-state functional magnetic resonance imaging.
Brain Structure and Function, 226:1067–1098.
Iordan, A. D., Cooke, K. A., Moored, K. D., Katz, B.,
Buschkuehl, M., Jaeggi, S. M., Jonides, J., Peltier,
S. J., Polk, T. A., and Reuter-Lorenz, P. A. (2018).
Aging and network properties: Stability over time and
links with learning during working memory training.
Frontiers in Aging Neuroscience, 9:419.
Jockwitz, C. and Caspers, S. (2021). Resting-state networks
in the course of aging—differential insights from stud-
ies across the lifespan vs. amongst the old. European
Journal of Physiology, 473:793–803.
Kitchenham, B. (2004). Procedures for performing system-
atic reviews. Keele University.
Lehmann, B. C. L., Henson, R. N., Geerligs, L., Cam-
CAN, and White, S. R. (2021). Characterising group-
level brain connectivity: A framework using bayesian
exponential random graph models. NeuroImage,
225:117480.
Li, W., Wang, M., Li, Y., Huang, Y., and Chen, X. (2016). A
novel brain network construction method for explor-
ing age-related functional reorganization. Computa-
tional Intelligence and Neuroscience, page 2429691.
Lv, H., Wang, Z., Tong, E., Williams, L., Zaharchuk, G.,
Zeineh, M., Goldstein-Piekarski, A., Ball, T., Liao,
C., and Wintermark, M. (2018). Resting-state func-
tional mri: Everything that nonexperts have always
wanted to know. American Journal of Neuroradiol-
ogy, 39(8):1390–1399.
Marek, S., Hwang, K., Foran, W., Hallquist, M. N., and
Luna, B. (2015). The contribution of network organi-
zation and integration to the development of cognitive
control. PLoS Biology, 13(12):e1002328.
McCreary, C. R., Salluzzi, M., Andersen, L. B., Gobbi,
D., Lauzon, L., Saad, F., Smith, E. E., and Frayne,
R. (2020). Calgary normative study: Design of a
prospective longitudinal study to characterise poten-
tial quantitative mr biomarkers of neurodegeneration
over the adult lifespan. BMJ Open, 10:e038120.
Meunier, D., Achard, S. ad Morcom, A., and Bullmore,
E. (2009). Age-related changes in modular organi-
Pilot Study of Distinct Graphs Models in Analysis of Brain Aging in Resting-State Functional Connectivity Networks
355

zation of human brain functional networks. NeuroIm-
age, 44(3):715–723.
Moretto, M., Silvestri, E., Zangrossi, A., Corbetta, M., and
Bertoldo, A. (2022). Unveiling whole-brain dynam-
ics in normal aging through hidden markov models.
Human Brain Mapping, 43:1129–1144.
Onoda, K. and Yamaguchi, S. (2013). Small-worldness
and modularity of the resting-state functional brain
network decrease with aging. Neuroscience Letters,
556:104–108.
Pedersen, R., Geerligs, L., Andersson, M., Gorbach, T.,
Avelar-Pereira, B., Wahlin, A., Rieckmann, A., Ny-
berg, L., and Salami, A. (2021). When functional blur-
ring becomes deleterious: Reduced system segrega-
tion is associated with less white matter integrity and
cognitive decline in aging. NeuroImage, 242:118449.
Rubinov, M. and Sporns, O. (2010). Complex network mea-
sures of brain connectivity: Uses and interpretations.
NeuroImage, 52:1059–1069.
Sala-Llonch, R., Junqu
´
e, C., Arenaza-Urquijo, E. M.,
Vidal-Pi
˜
neiro, D., Valls-Pedret, C., Palacios, E.,
Dom
`
enech, S., Salv
`
a, A., Bargall
´
o, N., and Bartr
´
es-
Faz, D. (2014). Changes in whole-brain functional
networks and memory performance in aging. Neuro-
biology of Aging, 35(10):2193–2202.
Sidhu, A. S. (2023). Decreasing Brain Functional Network
Segregation with Healthy Aging. University of Cal-
gary, Calgary, Canada.
Song, J., Birn, R. M., Boly, M., Meier, T. B., Nair,
V. A., Meyerand, M. E., and Prabhakaran, V. (2014).
Age-related reorganizational changes in modularity
and functional connectivity of human brain networks.
Brain Connectivity, 4(9):662–676.
Stumme, J., Jockwitz, C., Hoffstaedter, F., Amunts, K., and
S, C. (2020). Functional network reorganization in
older adults: graph-theoretical analyses of age, cogni-
tion and sex. Neuroimage, 214:116756.
Thomason, M. E., Brown, J. A., Dassanayake, M. T., Shas-
tri, R., Marusak, H. A., Hernandez-Andrade, E., and
Romero, R. (2014). Intrinsic functional brain archi-
tecture derived from graph theoretical analysis in the
human fetus. PLoS One, 9:e94423.
van den Heuvel, M. I., Turk, E., Manning, J. H., Hect, J.,
Hernandez-Andrade, E., Hassan, S. S., and Thoma-
son, M. E. (2018). Hubs in the human fetal brain
network. Developmental Cognitive Neuroscience,
30:108–115.
van den Heuvel, M. P., Lange, S. C., Zalesky, A., Seguin,
C., Yeo, B. T. T., and Schmidt, R. (2017). Pro-
portional thresholding in resting-state fmri functional
connectivity networks and consequences for patient-
control connectome studies: Issues and recommenda-
tions. NeuroImage, 152:437–449.
Varangis, E., Habeck, C. G., Razlighi, Q. R., and Stern, Y.
(2019). The effect of aging on resting state connectiv-
ity of predefined networks in the brain. Frontiers in
Aging Neuroscience, 11:e00234.
Wang, Q., Qi, L., He, C., Feng, H., and Xie, C. (2024).
Age- and gender-related dispersion of brain networks
across the lifespan. GeroScience, 46:1303–1318.
Wen, X., Zhang, H., Li, G., Liu, M., Yin, W., Lin, W., and
Shen, D. (2019). First-year development of modules
and hubs in infant brain functional networks. Neu-
roImage, 185:222–235.
Xiao, Y., Friederici, A. D., Margulies, D. S., and Brauer,
J. (2016). Longitudinal changes in resting-state fmri
from age 5 to age 6 years covary with language devel-
opment. NeuroImage, 128:116–124.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
356
