
First Results on Graph Similarity Search in Resting-State Functional
Connectivity Networks Using Spectral and Graph Edit Distances
M. A. G. Carvalho
1,2,3 a
and R. Frayne
2,3,4,5 b
1
School of Technology, University of Campinas, Brazil
2
Radiology and Clinical Neurosciences, Hotchkiss Brain Institute, Univ. of Calgary, Canada
3
Seaman Family MR Research Centre, Foothills Medical Centre, Calgary, Canada
4
Biomedical Engineering Graduate Program, University of Calgary, Canada
5
Calgary Image Processing and Analysis Centre, Foothills Medical Centre, Calgary, Canada
Keywords:
Brain Aging, rs-fMRI, Graph Modelling, Bold Signal, Graph Dissimilarity.
Abstract:
The application of graph theory in the modeling and analysis of brain networks has generated both new op-
portunities as well as new challenges in neuroscience. Resting state functional connectivity (RSFC) networks
studied with graphs is an important field of investigation because of the potential benefits in understanding
function in healthy individuals and identifying evidence of brain diseases and injury in patients. This work
is unique because it applies information retrieval techniques to create ranked lists from RSFC graph theory-
derived networks. In our analysis, we used a sample of whole-brain resting-state functional magnetic reso-
nance imaging (rs-fMRI) data obtained from Young (n = 10, age: 20.1 ± 2.1) and Old (n = 10, 65.6 ± 0.4)
sex-balanced groups drawn from a healthy, i.e., neurotypical, cohort. We estimated two well-known distance
metrics (graph edit distance and graph spectral distance) and by using information-retrieval graph ranking
methods achieved precision measures at the top-5 positions of ranked lists of up to 80%.
1 INTRODUCTION
Functional magnetic resonance imaging (fMRI) has
been used to understand human brain functions in
both healthy subjects and patients for over three
decades (Lv et al., 2018). Since the 2000s, resting-
state fMRI (rs-fMRI) data, acquired in the absence
of a task, has lead to the development of functional
connectivity (FC) measures. Resting state approaches
due to the relative simplicity of acquisition and con-
ceptual simplicity of analysis are frequently applied
to study healthy individuals and across patients with
a variety of neurological and psychiatric conditions.
Conditions like Alzheimer’s disease and Tourette’s
syndrome, for example, are associated with abnormal
alterations in connectivity between different brain re-
gions (Dai et al., 2019; Yang et al., 2023).
Among the many existing strategies to analyze rs-
fMRI data, graph theoretical approaches have been
applied due to their ability to investigate large, com-
plex networks. We denote a graph G = (V, E) by using
a
https://orcid.org/0000-0002-1941-6036
b
https://orcid.org/0000-0003-0358-1210
V to represent a set of nodes or vertices and E ⊆ V
2
to describe the set of connecting edges. Typically, the
analysis of RSFC networks is done by obtaining and
comparing measurements obtained from the graph G,
such as modularity, clustering coefficient, between-
ness centrality, global efficiency, and network degree
and density(Bullmore and Sporns, 2009). Graphs
have been used in modeling resting-state functional
connectivity (RSFC) networks by several research
groups (Wu et al., 2023; Wright et al., 2021; Hry-
bouski et al., 2021).
Graph similarity search methods, used com-
monly in information science, represents alternate ap-
proaches to analyze RSFC networks. They have not
been previously applied to study RSFC networks in
either healthy individuals or patients. Graph simi-
larity search seeks to retrieve relevant graphs from
a user-specified graph-structured query (Liang and
Zhao, 2017). Usually, distance or similarity metrics
are used to compute overall measures that account
for the underlying relationships amongst the graphs.
Ranked lists from the perspective of a graph query
can be calculated using these distance or similarity
Carvalho, M. A. G. and Frayne, R.
First Results on Graph Similarity Search in Resting-State Functional Connectivity Networks Using Spectral and Graph Edit Distances.
DOI: 10.5220/0013261400003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 357-362
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
357

measures.
We used two metrics to calculate distance in
graphs: 1) graph edit distance (GED), and 2) graph
spectral distance (GSD). GED estimates the dissimi-
larity measure between two graphs, G
1
and G
2
, that is
calculated by finding the minimum number of stan-
dard graph edit operations needed to transform G
1
into G
2
(Riesen, 2015). The standard set of edit
operations includes insertions and deletions of both
nodes and edges. GSD provides a second measure
of similarity between G
1
and G
2
. Typically, GSD
uses the eigenvectors and eigenvalues of the graph
adjacency or Laplacian matrix and computes the Eu-
clidean distance between the two graphs (Wilson and
Zhu, 2008). To our knowledge, graph similarity
search methods have not been applied to RSFC net-
works in healthy individuals or patients.
In this paper, we used graph similarity search ap-
proaches to assess RSFC whole-brain networks of
Young and Old healthy (neurotypical) individuals.
Ranked lists, based on GED and GSD, were used to
evaluate our graphs.
We examined the following research question:
Can GED or GSD be used as an efficient measure
of dissimilarity of RSFC networks of individuals in
different age groups? Section 2 presents our analysis
pipeline and describes the key methodological steps,
including how we modeled RSFC networks and the
calculated GED and GSD. Section 3 presents the ex-
perimental results and Section 4 summarizes our con-
clusions, study limitations and future work.
2 COMPUTING GRAPH
DISTANCES AND RANKED
LISTS
In this section, we describe the approach used to com-
pute network distances from a graph model represent-
ing RSFC networks. Our approach consists of three
main steps (outlined by the dotted squares in Figure
1). Each step is described in the following three Sub-
sections.
2.1 Dataset and FC Matrix
Data from the Calgary Normative Study (CNS) (Mc-
Creary et al., 2020) was used in this work. The CNS is
an ongoing study, begun in 2013, that focuses on col-
lecting quantitative MR data from healthy adults over
18 years. All MR data were acquired from individuals
residing in or near the Calgary, Alberta, Canada who
provided informed written consent. Data acquisition
was approved by the University of Calgary Conjoint
Health Research Ethics Board. The CNS acquires
several types of quantitative MR neuroimaging data
including rs-fMRI. We selected N = 20 individuals
from the CNS database. The demographics for the
Young and Old groups are shown in Table 1. Both
groups had a 50% : 50% female : male sex balance.
Table 1: Sex (Female, Male) and group (Young and Old)
demographics. Unpaired t-tests were used to determine sig-
nificance by sex and by group.
Sex Count Age(years) p-value
Female n = 10 44.3 ± 23.7 0.948
Male n = 10 43.6 ± 23.8
Cohort Count Age(years)
Young n = 10 20.1 ± 2.1 < 0.001
Old n = 10 65.6 ± 0.4
Total n = 20 44.0 ± 23.8
In the CNS study, rs-fMRI data were acquired
by measuring spatially localized fluctuations in the
blood oxygen level dependent (BOLD) signal. This
signal included noise and artifact from a variety of
sources. A processing pipeline that comprised sev-
eral steps was applied to extract the BOLD fluctu-
ations from this signal. The pipeline included the
analysis and preparation of rs-fMRI images, as de-
scribed in (Sidhu, 2023). Briefly the pipeline in-
cluded: skull striping using the Brain Extraction
Tool (BET), motion correction using the Motion
Correction FMRIB Linear Image Registration Tool
(MCFLIRT), interleaved slice-time correction, spatial
smoothing, temporal high-pass filtering, independent
component analysis (ICA) and functional-structural
registration. Structural and functional image prepro-
cessing was carried using publicily available soft-
wares FreeSurfer(Fischl, 2012) and FSL(Smith et al.,
2004), respectively. For each individual, a FC matrix
of size 200 × 200 was calculated. This matrix size
corresponded to segmenting the whole-brain into 200
anatomical regions using the Schaefer-Yeo cortical at-
las. After analyzing the quality of the rs-fMRI data, it
was decided to exclude graph nodes from the left and
right limbic networks because of signal loss result-
ing from MR susceptibility artifacts. Finally, Fisher’s
r–to–z transformation was applied to the Pearson cor-
relation values. An example FC matrix is shown in
Figure 2(a), where the colors are associated with the
Pearson correlation coefficient values.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
358

Figure 1: Analysis pipeline overview showing main steps (denoted by dotted boxes, from left to right): 1) creation of the
functional connectivity (FC) matrix, 2) modeling of the graph and 3) estimation and ranking of graph distance). CNS =
Calgary Normative Study(McCreary et al., 2020).
(a)
(b)
Figure 2: (a) Example of Pearson FC correlation matrix
and (b) its corresponding graph of one individual from the
CNS dataset. For visualization purposes, the graph in (b)
was built for a graph density equal to 5%.
2.2 Graph Model for RSFC Network
An undirected graph is an ordered pair G = (V, E)
with a node (or vertex) set V and an edge (or link)
set E connecting the nodes (Chung, 2019). An undi-
rected graph yields a symmetric connectivity matrix.
Each anatomical region resulting from the brain par-
cellation is a graph node u ∈ V and pairs of nodes
represent a connection. Each element of the FC ma-
trix represents the edge e ∈ E between a pair of nodes.
The associated r-to-z transformed Pearson correlation
value of the RSFC network is the edge weight. In
order to characterize a RSFC network as a “small-
world” network (where most nodes maintain only a
few direct connections) several strategies are com-
monly employed including assuming the entire net-
work is a graph, applying a thresholding operation,
creating a spanning tree and adopting a specific graph
density (Deery et al., 2023; Jockwitz and Caspers,
2021).
We chose to use the graph density modeling ap-
proach with an initial seed given by maximum span-
ning tree (MaxST). The reason for this choice is that
using MaxST ensures that we obtain a connected
graph, which is necessary for calculating the GSD.
Furthermore, MaxST iteratively selects the most rel-
evant edges in its construction. In this work, the
MaxST is derived from an undirected graph for each
participant, built with N nodes that matched the size
of FC matrix, and a number of edges E selected to
represent a pre-specified fraction of the graph den-
sity, i.e., a number of selected entries (or links) in the
FC matrix. We used only positive Pearson correlation
values (z ⩾ 0) and set the main diagonal of the FC
matrix to zero to exclude self-connections.
Based on literature findings, we considered using
graph densities ranging from [1%, 25%] (Marek et al.,
2015) to [22%, 40%](Grady et al., 2016). To better
fine tune the graph density range, we considered two
criteria: First, we used the modularity metric (M) as a
reference metric because 94% of studies demonstrate
decrease in M with age(Deery et al., 2023). Modu-
larity measures the topological organization of whole
brain FC networks in a set of groups, where it is possi-
First Results on Graph Similarity Search in Resting-State Functional Connectivity Networks Using Spectral and Graph Edit Distances
359

ble to distinguished dense internal (intra-module) and
sparse external (inter-module) connectivity. Second,
values of M ⩾ 0.3 are generally associated with non-
random module structure and, thus, are thought to re-
flect brain topological organization. After this anal-
ysis we decided to use a graph density ranging from
[1%, 10%].
Figure 2(b) shows an example graph, correspond-
ing to a percentage of edges of a complete graph con-
structed from the FC correlation matrix of Figure 2(a).
2.3 Graph Distance
Two distance measurements were considered:
1) Graph Edit Distance (GED) is a dissimilarity mea-
sure from the number as well as the strength of the op-
erations that have to be applied to transform a source
graph (G
1
) into a target graph (G
2
)(Riesen, 2015).
GED is defined by:
GED(G
1
, G
2
) = min
∑
i
c(e
i
) (1)
where c(e
i
) denotes the cost of the i
th
edit opera-
tion. In this work, considering that graphs (RSFC net-
works) have the same dimension and the same label-
ing of nodes anatomical regions), edit operations are
limited to the deletion and insertion of edges, with the
cost of each operation being equal to 1. In addition, a
variant of GED that considers only the weights of the
added or removed edges was also calculated.
2) Graph Spectral Distance (GSD) is obtained from
the spectrum of the graph derived from its adjacency
matrix representation using eigenvalue decomposi-
tion. The graph spectrum is the set of ordered eigen-
values s
i
= {λ
i
1
, λ
i
2
, ..., λ
i
|V|
}, where i refers to the
graph label, | V | represents the size or the number
of nodes of the matrix G corresponding to the RSFC
network. GSD is defined by:
GSD(G
1
, G
2
) =
r
∑
i
(s
1
− s
2
)
2
=
=
q
(λ
1
1
− λ
2
1
)
2
+ ... +(λ
1
|V|
− λ
2
|V|
)
2
(2)
where the subscript denotes the number of the eigen-
value and the superscript, refers to the graphs 1 and
2. In this work, the GSD is computed by using the
Laplacian matrix L = D − A, where D is the diago-
nal degree matrix and A corresponds to the graph ad-
jacency matrix. Additionally, GSD was also calcu-
lated from the Euclidean distance between the second
smallest eigenvector of each RSFC matrix, known as
the Fiedler vector (Wilson and Zhu, 2008).
2.4 Ranked Lists and Evaluation
We adapted the notation defined by (Pedronette et al.,
2016). Let C = {G
1
, G
2
, ..., G
n
} be a collection of
graphs, where n =| C | is the size of C. Based on
the distance measure, d(·, ·), a ranked list τ
G
q
can be
computed as a permutation of the collection C in re-
sponse to a query graph G
q
. If G
i
is ranked before G
j
in the ranked list of G
q
then d(G
q
, G
i
) ≤ d(G
q
, G
j
).
Every graph G
q
∈ C can produce a ranked list. There-
fore, a set of ranked lists R = {τ
G
1
, τ
G
2
, ..., τ
G
n
} can
be obtained. In order to evaluate the list of ranked
lists, we compute the precision p (the fraction of rele-
vant list entries among the retrieved list) at position k
(i.e., p@k). In other words, p@k is the fraction of the
number of relevant items in the first k positions of the
ranked list.
3 RESULTS AND DISCUSSIONS
Figure 3 plots the p@5 to p@10 values as a function
of graph density for GED and GSD computed accord-
ing to a graph query for identifying the Young group.
The most relevant information (around 80% of preci-
sion) were obtained at the top-5 positions of ranked
lists, i.e., at p@5. They are compatible with what
the information retrieval literature suggests regarding
the value of k << n where n =| C | is the number of
graphs(Pedronette et al., 2016). An 80% value for
p@5 means that for the obtained ranked lists, four of
the first five items were relevant and correctly identi-
fied a RSFC network from an individual in the Young
group.
In this type of study it is also possible to observe
how representative a given RSFC network is for either
the Young or Old groups. The p@5 sequence across
the ten ranked lists for GED was (80%, 80%, 80%,
80%, 60%, 100%, 100%, 80%, 80%, 60%), where
each element corresponds to one of the ten individuals
in the Young group. These values were obtained for a
graph density of 1%. Some networks reached 100%
precision while others only achieved 60%. Lower pre-
cision values may serve as indicators or biomarkers
that should result in further investigations for a spe-
cific individual. Finally, the analysis of ranked lists
with higher precision values can contribute to defin-
ing graph density values or range of values that are
most appropriate to represent a RSFC network.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
360
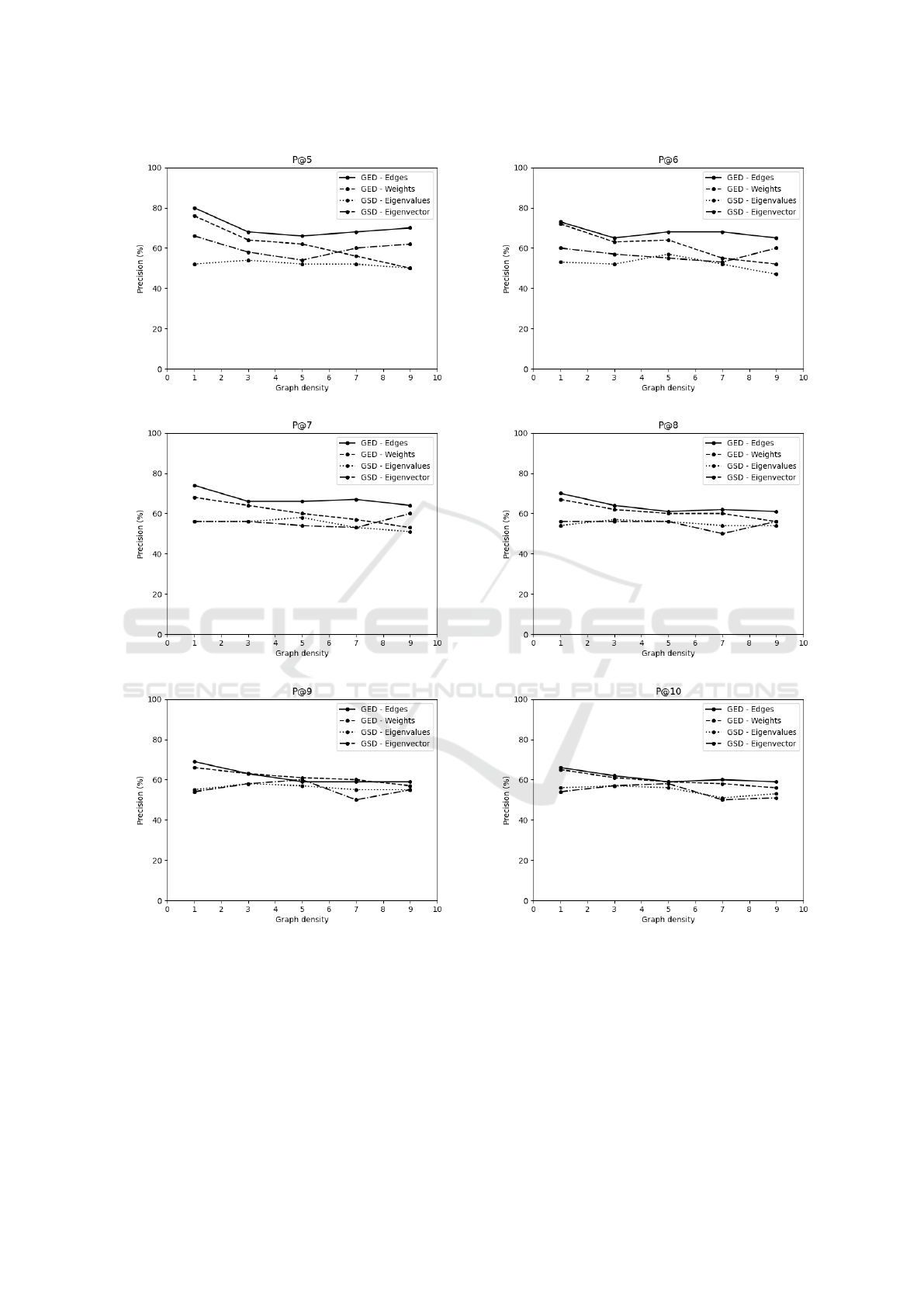
Figure 3: Average precision p@k (k = 5 to 10) to rank individuals in the Young group.
4 CONCLUSIONS
In the current study we have shown that techniques
used in the area of graph similarity search can be ap-
plied to analyze RSFC networks. The use of ranked
lists is useful in the information retrieval processes,
such as identifying Young or Older individuals as in
this work. As well, ranked lists may being applied for
other types of tasks, such as the clustering of RSFC
networks of individuals with different types of dis-
eases. The best precision values obtained in this work
were around 80% and suggests that there is a possibil-
ity of increase through the use of re-ranking and rank-
ing aggregation techniques (Pedronette et al., 2016).
First Results on Graph Similarity Search in Resting-State Functional Connectivity Networks Using Spectral and Graph Edit Distances
361

We intend to expand this study by analyzing the entire
set of images collected by the CNS, considering the
inclusion and exclusion criteria indicated in (Sidhu,
2023), and observing the quality of the scans in each
of the functional networks in the preprocessing stage.
Further studies should extend these whole-brain re-
sults and individually examine sensory and associa-
tive functional networks that are consistently reported
in the literature: Visual, Sensorimotor, Dorsal Atten-
tion, Ventral Attention, Limbic, Frontoparietal, and
Default Mode.
ACKNOWLEDGEMENTS
This study was financed in part by the Coordination of
Improvement of Higher Education Personnel - Brazil
(CAPES) - Finance Code 001. Marco Carvalho wish
to express their gratitude to the S
˜
ao Paulo Research
Foundation/Fundac¸
˜
ao de Amparo
`
a Pesquisa do Es-
tado de S
˜
ao Paulo (FAPESP grant 2023/02302-6). We
also acknowledge the assistance of Kau
ˆ
e TN Duarte,
Abhi S Sidhu, and Cherly R McCreary, from Univer-
sity of Calgary.
REFERENCES
Bullmore, E. and Sporns, O. (2009). Complex brain net-
works: graph theoretical analysis of structural and
functional systems. Nature Reviews Neuroscience,
10:186–198.
Chung, M. K. (2019). Brain Network Analysis. Cambridge
University Press, United Kingdom.
Dai, Z., Lin, Q., Li, T., Wang, X., Yuan, H., Yu, X., He, Y.,
and Wang, H. (2019). Disrupted structural and func-
tional brain networks in Alzheimer’s disease. Neuro-
biology of Aging, 75:71–82.
Deery, H. A., Paolo, R. D., Moran, C., Egan, G. F., and
Jamadar, S. D. (2023). The older adult brain is less
modular, more integrated, and less efficient at rest:
A systematic review of large-scale resting-state func-
tional brain networks in aging. Psychophysiology,
60:e14159.
Fischl, B. (2012). Freesurfer. Neuroimage, 62(2):774.
Grady, C., Sarraf, S., Saverino, C., and Campbell, K.
(2016). Age differences in the functional interac-
tions among the default, frontoparietal control, and
dorsal attention networks. Neurobiology of Aging,
41:159–172.
Hrybouski, S., Cribben, I., McGonigle, J., Olsen, F., Carter,
R., Seres, P., Madan, C. R., and Malykhin, N. V.
(2021). Investigating the effects of healthy cogni-
tive aging on brain functional connectivity using 4.7
T resting-state functional magnetic resonance imag-
ing. Brain Structure and Function, 226:1067–1098.
Jockwitz, C. and Caspers, S. (2021). Resting-state network
in the course of aging - differential insights from stud-
ies across the lifespan vs. amongst the old. European
Journal of Physiology, 473:793–803.
Liang, Y. and Zhao, P. (2017). Similarity search in graph
databases: A multi-layered indexing approach. In
2017 IEEE 33rd International Conference on Data
Engineering (ICDE), pages 783–794.
Lv, X. H., Wang, X. Z., Tong, X. E., Williams, X. L., Za-
harchuk, X. G., Zeineh, X. M., Goldstein-Piekarski,
X. A., Ball, X. T., Liao, X. C., and Wintermark, X. M.
(2018). Resting-state functional MRI: Everything that
nonexperts have always wanted to know. AJNR Am J
Neuroradiol, 39:1390–1399.
Marek, S., Hwang, K., Foran, W., Hallquist, M. N., and
Luna, B. (2015). The contribution of network organi-
zation and integration to the development of cognitive
control. PLoS Biology, 13(12):e1002328.
McCreary, C. R., Salluzzi, M., Andersen, L. B., Gobbi,
D., Lauzon, L., Saad, F., Smith, E. E., and Frayne,
R. (2020). Calgary normative study: Design of a
prospective longitudinal study to characterise poten-
tial quantitative MR biomarkers of neurodegeneration
over the adult lifespan. BMJ Open, 10:e038120.
Pedronette, D. C. G., Almeida, J., and Torres, R. S. (2016).
A graph-based ranked-list model for unsupervised dis-
tance learning on shape retrieval. Pattern Recognition
Letters, 83:357–367.
Riesen, K. (2015). Structural Pattern Recognition with
Graph Edit Distance. Springer, Switzerland.
Sidhu, A. S. (2023). Decreasing Brain Functional Network
Segregation with Healthy Aging. MSc thesis, Biomed-
ical Engineering, University of Calgary, Calgary, AB,
Canada.
Smith, S. M., Jenkinson, M., Woolrich, M. W., Beckmann,
C. F., Behrens, T. E. J., Johansen-Berg, H., Bannis-
ter, P. R., Luca, M. D., Drobnjak, I., Flitney, D. E.,
Niazy, R. K., Saunders, J., Vickers, J., Zhang, Y., Ste-
fano, N. D., Brady, J. M., and Matthews, P. M. (2004).
Advances in functional and structural mr image anal-
ysis and implementation as fsl. Neuroimage, 23:Suppl
1:S208–19.
Wilson, R. C. and Zhu, P. (2008). A study of graph spectra
for comparing graphs and trees. Pattern Recognition,
41(12):2833–2841.
Wright, L. M., Marco, M. D., and Venneri, A. (2021). A
graph theory approach to clarifying aging and disease
related changes in cognitive networks. Frontiers in
Aging Neuroscience, 13:676618.
Wu, K., Jelfs, B., Mahmoud, S. S., Neville, K., and Fang,
J. Q. (2023). Tracking functional network connectiv-
ity dynamics in the elderly. Frontiers in Neuroscience,
17:1146264.
Yang, Y., Yang, H., Yu, C., Ni, F., Yu, T., and Luo, R.
(2023). Alterations in the topological organization of
the default-mode network in Tourette syndrome. BMC
Neurology, 23:390.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
362
