
Mitigating Algorithmic Bias in Prostate Cancer Risk Stratification with
Responsible Artificial Intelligence and Machine Learning
Meghana Kshirsagar
1,2,4 a
, Mihir Sontakke
1
, Gauri Vaidya
1,2,4 b
, Ahmad Alkhan
3,4 c
,
Aideen Killeen
1,2
and Conor Ryan
1,2,4 d
1
Department of Computer Science and Information Systems, University of Limerick, Ireland
2
Lero the Research Ireland Centre for Software, Ireland
3
School of Medicine, University of Limerick, Ireland
4
Limerick Digital Cancer Research Centre, Ireland
Keywords:
Artificial Intelligence, Prostate Cancer, Algorithmic Bias, Image Triage, Deep Learning, Machine Learning.
Abstract:
Prostate cancer (PCa) is the second most prevalent cancer among men worldwide, the majority affecting those
over the age of 65. The Gleason Score (GS) remains the gold standard for diagnosing clinically significant
prostate cancer (csPCa); however, traditional biopsy can lead to patient discomfort. Algorithmic bias in med-
ical diagnostic models remains a critical challenge, impacting model reliability and generalizability across
diverse patient populations. This study explores the potential of Machine Learning (ML) models—Logistic
Regression (LR) and multiple DL models—as non-invasive alternatives for predicting the GS using Prostate
Imaging Cancer AI challenge dataset . To the best of our knowledge, this is the first attempt to use two modal-
ities with this dataset for risk stratification. We developed a LR model, excluding biopsy-derived features like
GS, to predict clinically significant prostate cancer, alongside an image triage approach with convolutional
neural networks to reduce biases in the ML workflow. Preliminary results from LR and ResNet50, showed
test accuracies of 69.79% and 60%, respectively. These findings demonstrate the potential for explainable,
trustworthy, and responsible risk stratification enhancing the robustness and generalizability of the prostate
cancer risk stratification model.
1 INTRODUCTION
Prostate cancer (PCa) resulted in approximately 1.5
million cases every year globally, out of which
397,000 had a fatal outcome in 2022 (Wang et al.,
2022). With an increased number of aging popula-
tion demographics in Organization for Economic Co-
operation and Development countries, prostate can-
cer are supposed to rise in the coming future (oec, ).
Traditional PCa diagnosis typically involves a combi-
nation of clinical evaluations such as the digital rec-
tal examination (DRE), serum prostate-specific anti-
gen (PSA) testing, and the gold standard of transrec-
tal ultrasound-guided prostate biopsy. PSA testing,
though widely used, has limitations, as it can yield
false positives or negatives, leading to unnecessary
a
https://orcid.org/0000-0002-8182-2465
b
https://orcid.org/0000-0002-9699-522X
c
https://orcid.org/0000-0002-5130-9767
d
https://orcid.org/0000-0002-7002-5815
biopsies or missed diagnoses. The biopsy itself, while
accurate, is invasive and carries risks like infection,
bleeding, and pain. Moreover, biopsy results may not
always provide a clear picture of the cancer’s aggres-
siveness or extent (Mottet et al., 2017).
MRI-scanned images provide a clear picture of
the prostate and nearby areas, allowing the identifi-
cation of PCa spread and identifying high-risk pa-
tients. Multiparametric magnetic resonance imaging
(mpMRI) is increasingly utilised globally as a non-
invasive tool to detect, localise, and stage PCa, allow-
ing prostate biopsy planning (Pecoraro et al., 2021).
As mpMRI adoption grows, it offers a promising ap-
proach to streamline prostate cancer diagnosis and
treatment decisions, reducing patient discomfort and
overdiagnosis.
Deep learning (DL) methodologies, notably con-
volutional neural networks (CNNs), have become in-
tegral in artificial intelligence for computer vision
tasks. These CNNs assist physicians by accelerating
tumor detection processes while ensuring high perfor-
Kshirsagar, M., Sontakke, M., Vaidya, G., Alkhan, A., Killeen, A. and Ryan, C.
Mitigating Algorithmic Bias in Prostate Cancer Risk Stratification with Responsible Artificial Intelligence and Machine Learning.
DOI: 10.5220/0013262600003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 3, pages 1085-1092
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
1085

mance and accuracy.
The application of deep learning has the poten-
tial to significantly enhance prostate cancer (PCa) de-
tection rates, leading to more accurate and expedited
diagnoses. This improvement can help prevent both
over-diagnosis and under-diagnosis. By diminishing
the reliance on costly diagnostic tests and invasive
biopsies, deep learning also offers a means to reduce
healthcare expenditures.
Interpreting MRI images presents substantial
challenges due to their inherent complexity (He et al.,
2023). This complexity contributes to considerable
intra-observer and inter-observer variability in image
readings, as documented in various studies (Pellicer-
Valero et al., 2022; Brembilla et al., 2020). In this
context, deep learning offers a promising solution by
streamlining the interpretation process, enhancing the
quality of image analysis, and reducing the potential
for overtreatment.
In this article, we aim to address several types
of bias through our image triage approach, including
confirmation, sample, demographic, equipment, and
protocol bias. Confirmation bias is mitigated by mix-
ing slices from both positive and negative classes, pre-
venting the model from being biased toward a specific
class. Sample bias is reduced by including a large
number of slices, ensuring diverse data representa-
tion. Demographic bias is addressed as the dataset
encompasses a cohort with ages ranging from 35 to
92 years. Finally, equipment and protocol bias is min-
imized by incorporating slices obtained from different
MRI sequencing types, ensuring a broader and more
generalized representation of the data.
2 RELATED WORKS
This section highlights the latest advancements in the
field of risk assessment in prostate cancer using ma-
chine learning (ML) and artificial intelligence (AI).
The study by Amirhossein Jalali et al. (Jalali et al.,
2023) focuses on developing and validating a risk cal-
culator to assist in determining whether a prostate
biopsy is necessary. The study was conducted at a
rapid access prostate cancer clinic, where data from
3,531 men who were referred for a suspected prostate
cancer diagnosis were collected. These patients were
primarily referred due to elevated PSA levels, abnor-
mal DRE findings, or a combination of both. The co-
hort included a wide range of men, from those at low
risk of prostate cancer to those at high risk, providing
a comprehensive dataset for developing and validat-
ing the risk calculator. Data collected included demo-
graphic information, clinical variables (e.g., age, PSA
levels, DRE results, prostate volume), and biopsy out-
comes. The biopsy results were used as the gold
standard to determine the presence of prostate cancer,
which was then categorized into clinically significant
and insignificant cancers based on established crite-
ria.
The authors employed logistic regression analy-
sis to identify key variables associated with the likeli-
hood of a positive prostate biopsy. Variables consid-
ered included PSA levels, DRE findings, prostate vol-
ume, and patient age. These variables were chosen
based on their established association with prostate
cancer risk and availability in routine clinical prac-
tice. The risk calculator model was developed using
a subset of the data and then validated on the remain-
ing cohort. The model provided a risk score for each
patient, indicating the probability of having a posi-
tive biopsy. This score could then be used by clini-
cians to make more informed decisions about whether
to proceed with a biopsy. The performance of the
risk calculator was assessed using several metrics, in-
cluding the area under the receiver operating char-
acteristic curve (AUC-ROC), sensitivity, specificity,
and calibration. The AUC-ROC is a measure of the
model’s ability to discriminate between patients with
and without prostate cancer, with a value of 1 indicat-
ing perfect discrimination and 0.5 indicating no dis-
crimination.
One of the key advantages of the risk calculator
is its ability to integrate multiple clinical variables
into a single risk score, offering a more comprehen-
sive assessment than relying on PSA levels or DRE
findings alone. There were several limitations of the
study. First, the study cohort was derived from a
single institution, which may limit the generalizabil-
ity of the findings to other populations or settings.
Second, the model was developed using retrospective
data, and prospective validation in a broader popula-
tion is needed to confirm its utility. Another limitation
is the reliance on biopsy results as the gold standard
for prostate cancer diagnosis. Biopsy itself is not a
perfect test, and some cancers may be missed, partic-
ularly those that are small or located in difficult-to-
sample areas of the prostate.
The authors in (Jalali et al., 2020) focus on im-
proving the accuracy of prostate cancer detection by
incorporating inflammatory serum biomarkers into
existing risk calculators and evaluate their associa-
tion with prostate cancer risk. These biomarkers were
then integrated into a modified risk calculator, along-
side traditional factors like PSA levels and patient de-
mographics. This integration allowed for enhanced
distinction between high-risk individuals and had su-
perior performance compared to the traditional risk
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1086
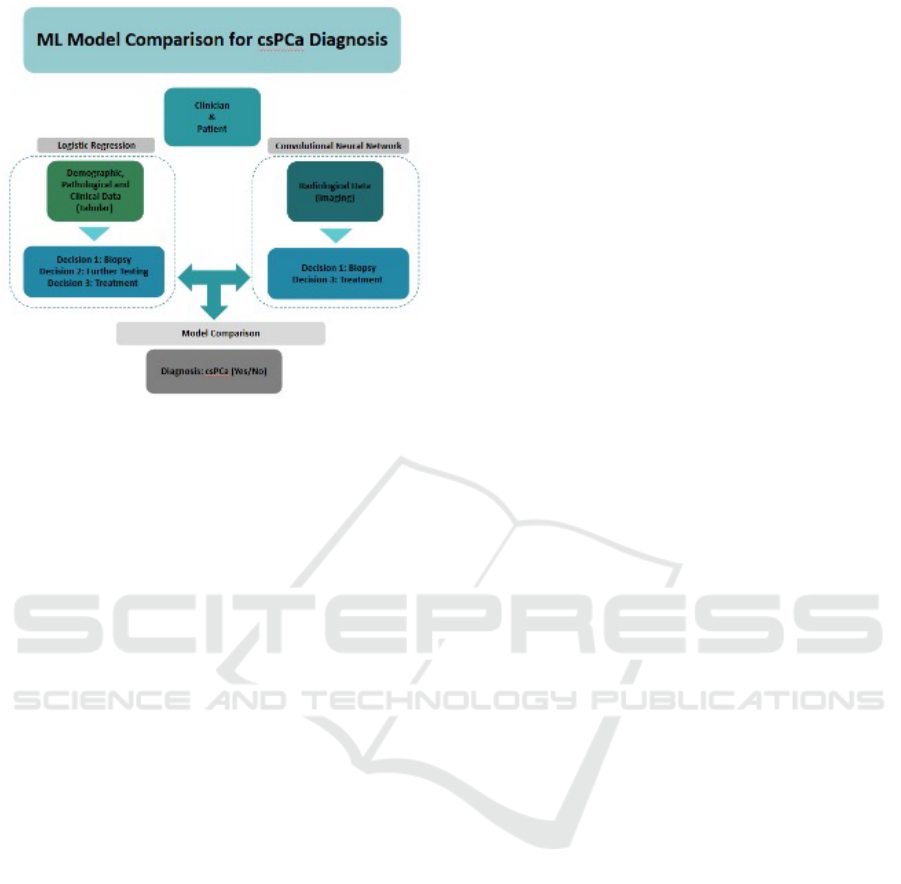
Figure 1: Classification methodologies for csPCa using LR
and CNN.
assessment tools. The authors concluded that incor-
porating inflammatory biomarkers has the promising
potential to refine prostate cancer risk stratification.
The biomarkers were integrated into a unified model
to assess their combined predictive power.
The paper by (Jalali et al., 2020) focuses on im-
proving the prediction of progression risk in patients
with castration-resistant prostate cancer (CRPC)
through the use of deep learning models that integrate
multiple types of data. The researchers employed a
multimodal deep learning model trained on data col-
lected from multiple centers. This model integrates
tabular clinical data, imaging features, and genomic
information to predict the risk of disease progression
in CRPC patients. The study demonstrated that the
integration of multimodal data significantly improves
the predictive accuracy of disease progression com-
pared to models that rely on a single data type. This
approach allows for a more personalized and precise
risk assessment for patients with CRPC.
3 METHODOLOGY
In this study, we employed two machine learn-
ing models to classify patients into binary cate-
gories: csPCa (Prostate Cancer) and non-csPCa (non-
Prostate Cancer). The first model, logistic regres-
sion, uses patient characteristics—specifically age,
PSA levels, PSAD (prostate-specific antigen density),
and prostate volume—while excluding the Gleason
score, a feature typically derived from biopsy analysis
(Hooshmand, 2021).
For the deep learning (DL) approach, we trained
a basic Convolutional Neural Network (CNN) (Jiang
et al., 2023) using the ResNet50 model on our im-
age triage strategy. In this method, slices (2-D cross-
sectional images within a plane) from both classes
across all patients were mixed. Each patient’s imag-
ing included captures from five different MRI se-
quences, giving a total of approximately 1422×5×25
images, based on the number of patients, MRI se-
quences, and average slices per sequence.
Both models output a binary decision, indicating
the presence or absence of prostate cancer. Logis-
tic regression is selected due to its clear separation
between dependent and independent variables, and it
outputs a predicted probability score based on the ob-
servation, which can be manually tuned to cater to
different use cases. CNNs are employed for their pro-
ficiency in capturing spatial hierarchies within image
data, thereby enabling accurate classification in the
context of medical imaging.
3.1 Study Design
This study was a retrospective observational cohort
study using publicly available real-world data, with
the effect size estimated based on prior research. The
ground truth for the machine learning models was
established through histopathology-confirmed diag-
noses of prostate cancer. Multiple data modalities
were used, with tabular data analysed using logistic
regression for binary classification, while MRI image
data were processed using convolutional neural net-
works (CNN). Model alignment was compared using
agreement statistics and confusion matrices. Figure 1
describes the ML model comparison methodology.
3.2 Study Population
The study population consisted of 1422 men between
35 and 92 years, including 1014 cases with benign
tissue or indolent PCa and 408 cases diagnosed with
csPCa. Patient inclusion required an abnormal rec-
tal exam, a PSA ≥ 3 ng/mL, or both. Patient exclu-
sion included a history of prostate-specific treatment.
Patients underwent MRI-targeted biopsy, transrectal
ultrasound-guided biopsies, a combination of both, or
radical prostatectomy for confirmatory PCa diagno-
sis. Patients with confirmed diagnoses ranged from
low to high-risk, stratified by Gleason score. The data
did not include clinical staging.
3.3 Data Repository
Data were acquired from the PI-CAI (Prostate Imag-
ing: Cancer AI) challenge; specifically, the pub-
Mitigating Algorithmic Bias in Prostate Cancer Risk Stratification with Responsible Artificial Intelligence and Machine Learning
1087

lic training and development dataset (Saha et al.,
2024). All data were fully anonymised and made
available under a non-commercial CC BY-NC 4.0 li-
cense. The dataset included demographic and clini-
cal data, histopathology results, and MRI scans from
three centres (11 sites) collected between 2011 and
2021.
3.3.1 Tabular Data
Clinical, demographic, and histopathological data in-
cluded patient age, PSA level, prostate volume, PSA
density, Gleason grade, ISUP grade, type of biopsy
procedure, and centre. The data does not differentiate
between biopsy sample types.
3.3.2 Imaging Data
Table 1: Details of MRI Sequences.
MRI Sequence Type Description
t2w Axial T2-weighted Imaging
adc Apparent Diffusion Coefficient map
hbv Diffusion-Weighted Imaging or DWI
cor Coronal T2-weighted Imaging
sag Sagittal T2-weighted Imaging
This dataset consists of five MRI-scanned image
types, as described in Table 1. This amounts to a
total of 74,050 images for model training, in the ra-
tio of 28.69% csPCa and 71.31% non-csPCa. All pa-
tient exams included bpMRI scans, axial T2-weighted
imaging (T2W), axial high b-value (≥ 1000 s/mm²)
diffusion-weighted imaging (DWI), and axial appar-
ent diffusion coefficient maps (ADC). In addition,
mpMRI scans were available for 1422 patients with
additional sagittal and coronal T2W scans. Scans
were acquired using Siemens Healthineers or Philips
Medical Systems-based scanners with surface coils.
Notably, no patient case contained dynamic contrast-
enhanced (DCE) sequences. Data also included basic
acquisition variables (scanner manufacturer, scanner
model name, diffusion b-value).
3.4 Data Pre-Processing and Cleaning
3.4.1 Tabular
Data cleaning and pre-processing followed best prac-
tice guidelines. Missing values were imputed, cate-
gorical variables were encoded using one-hot encod-
ing, and selected categories were summed and nor-
malised using a min-max scaler.
3.4.2 Imaging
The dataset consisted of images from 1422 patients,
each with slices captured from five different angles,
(a) (b)
Figure 2: Sample of an T2W image for PCa: (a) positive
and (b) negative class.
resulting in a variable number of slices per patient
(19 to 31 slices). The images were organised into
two classes: the ”yes” folder for clinically significant
prostate cancer (csPCa), which contained 21,250 im-
ages, and the ”no” folder for non-significant prostate
cancer (non-csPCa), containing 52,800 images. In to-
tal, 74,050 images were used for training and evalua-
tion of the model.
In the pre-processing pipeline, the three-
dimensional MRI images (.mha files) were converted
into two-dimensional .jpg images or slices from each
.mha image using the SimpleITK library, a widely
used tool for medical image processing (Biederer
et al., 2016).
Image slices were resized to a uniform dimen-
sion of (256 x 256) and converted into NumPy arrays.
Corresponding labels were encoded and organised to
align with the processed images for subsequent anal-
ysis. Matplotlib, PIL, NumPy, and OpenCV were em-
ployed to resize and process the tasks.
The machine learning process consisted of three
experiments: Experiments 1 and 2 used T2-weighted
(T2W) images, while Experiment 3 included all im-
age types. Each experiment followed an 85:15 split
ratio for training and testing datasets, resulting in
Xtrain and Ytrain for training and Xtest and Ytest for
testing, with labels csPCa.
3.5 Exploratory Data Analysis
Image files were converted to .jpeg via a DICOM con-
verter. The following Figure 2 shows prostate cancer
positive (a) and negative (b) T2W images.
Exploratory Data Analysis was carried out to sum-
marise the dataset, including missing values, sorting
data types, measuring class imbalance, and under-
standing data distributions.
Figure 3 illustrate the class imbalance present
within the tabular dataset, highlighting discrepancies
in the representation of different PCa categories. The
class imbalance for the combined MRI images was
analysed, as shown in Figure 4. The class imbalance
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1088
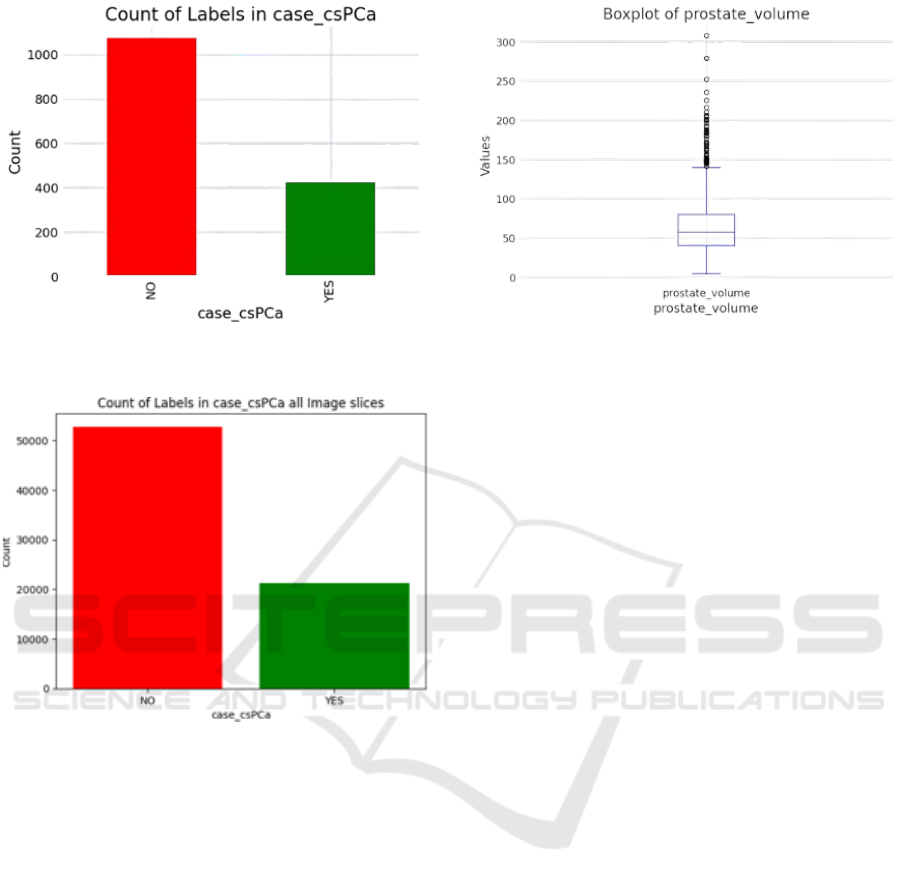
Figure 3: Bar plot representing data distribution across the
two classes for tabular dataset.
Figure 4: Bar plot representing the classes within the dataset
within csPCA.
in the image labels posed a potential risk to model per-
formance. While data augmentation techniques, such
as horizontal flipping, could address this imbalance,
they also risk distorting critical medical features. As a
result, to address class imbalance, our approach mixes
all images from both classes to train the CNN on a
rich and diverse feature set containing patterns with
subtle variations. The distribution of prostate volume
in Figure 5 aligns with trends observed in other real-
world datasets. Adjusting volume-related image dif-
ferences and employing normalisation techniques in
training can help mitigate the risk of overfitting to
extreme cases, ultimately enhancing the utility of AI
tools in real-world settings.
3.6 Machine Learning Models
The dataset was divided into independent (X) and de-
pendent (Y) variables and split into training and test-
ing sets with an 80:20 ratio. To address class im-
Figure 5: Boxplot of Prostate Volume. The median value of
the prostate volume is 57 cubic centimetres (cc).
balance, SMOTE was applied to generate synthetic
data for the minority class in the binary csPCa label.
Once the data were split, class balance was achieved,
and model training proceeded. A logistic regression
model was employed to predict whether the patient
has clinically significant prostate cancer.
3.6.1 Model Training
For the deep learning (DL) approach, we trained
several models, including a standard Inception v3
and ResNet50 model, using an image triage strat-
egy. Model training was performed using Python
with TensorFlow, Sklearn, and SimpleITK libraries
on NVIDIA A100 80GB PCIe to accelerate compu-
tation.
3.6.2 Model Building
Transfer learning was used to train and optimise four
CNN models based on Inception v3 (Szegedy et al.,
2016) and ResNet 50 (He et al., 2016) architectures.
Both models employed pre-trained ImageNet weights
and were fine-tuned to extract deep feature represen-
tations specific to the MRI data. Each model was
adapted to binary classification tasks, and the final
layers were modified accordingly.
3.6.3 Data Pipeline
Data augmentation was applied to the training set us-
ing the Image Data Generator, including re-scaling
the pixel values by dividing by 255. Both test and
train data were re-scaled. Horizontal flips were ex-
cluded to preserve the orientation of medical images.
Models were trained with batch sizes of 64 across 40
epochs with a data split of training (64%), validation
(16%), and test (20%), respectively. The experiments
were conducted using a full image dataset.
Mitigating Algorithmic Bias in Prostate Cancer Risk Stratification with Responsible Artificial Intelligence and Machine Learning
1089
3.6.4 Model Training
All models were trained with an Adam optimiser
(learning rate = 0.0001), binary cross-entropy loss
function, employing early stopping over 10 epochs to
prevent overfitting.
Setup 1: Inception v3
A pre-trained Inception v3 model from TensorFlow
with ImageNet weights was employed, utilising trans-
fer learning. Layers at both ends of the network were
excluded to maintain the original feature extraction
capability, with the remaining layers frozen to prevent
updates during training. The output was flattened, fol-
lowed by the application of a dense layer with a single
neuron and a sigmoid function for binary classifica-
tion. This configuration enabled the model to effec-
tively classify data into two categories with minimal
modification to the original Inception architecture.
Setup 2: Inception v3 with Additional Layer Ar-
chitecture
The second model builds on the Experiment 1 model
setup by incorporating additional layers with three
dense and three dropout layers following the Incep-
tion v3 output.
Setup 3: ResNet 50
The third model employs the ResNet 50 pre-trained
model, using a similar approach to the Inception v3
model from Experiment 1. The layers of ResNet 50
are frozen to retain the pre-learned features, while the
final layers are adapted for the binary classification
task.
Setup 4: ResNet 50 with Additional Layer Archi-
tecture
The fourth model builds upon the ResNet 50 archi-
tecture used in Experiment 3, adding similar modifi-
cations made in the second model from Experiment
1. These modifications include the addition of layers
after the output of the pre-trained ResNet model.
4 RESULTS
This section discusses the results from the tabular and
image modalities.
4.1 Training Results
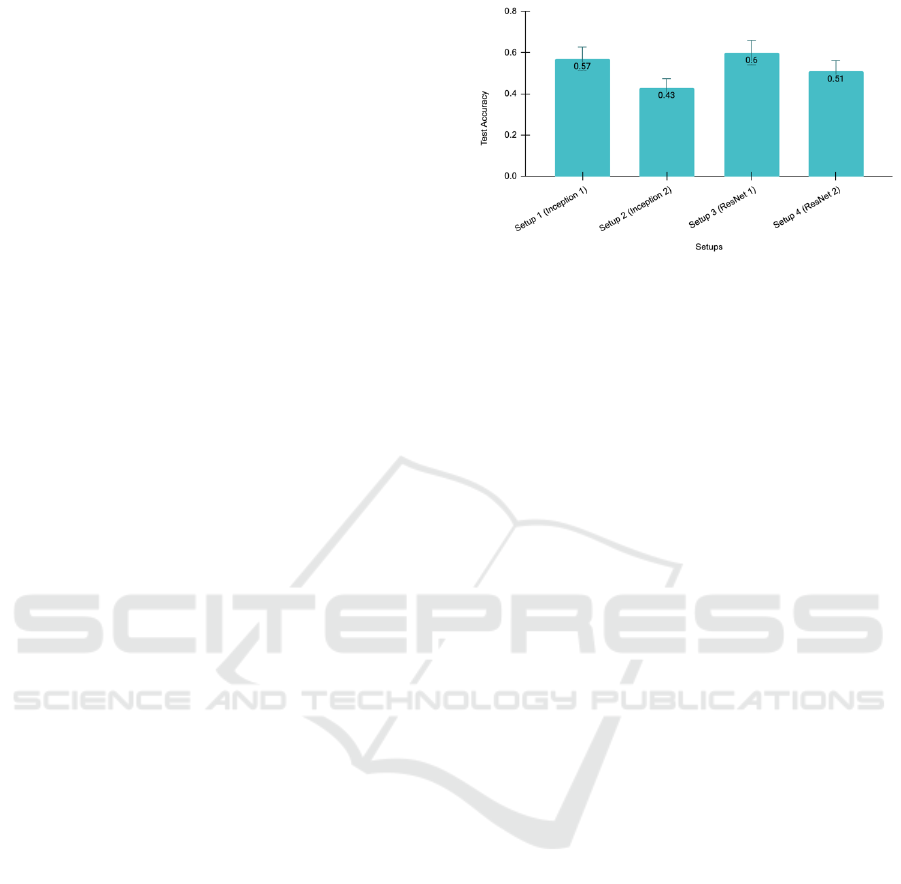
The LR model achieved a test accuracy of 69.79%.
On the other hand, the test accuracies of the multiple
setups with the image modality are shown in Figure 6.
The ResNet1 model (Setup 3) outperforms all the se-
tups. Therefore, ResNet1 was selected as the final
model.
Figure 6: Comparative analysis between the experimental
setups.
4.2 Confusion Matrix
The alignment of the model’s predictions with actual
outcomes, assessed through concordance percentage
and as illustrated by the confusion matrix, highlights
the model’s performance in classifying csPCa and
non-csPCa cases. The confusion matrix (Figure 7)
indicates that the model has a high proportion of
True Negatives (TN) (49.9%), suggesting it effec-
tively identifies non-csPCa cases, while the True Pos-
itive (TP) rate (20.2%) shows reasonable detection of
csPCa, and the False Positive (FP) rate (49.9%) is sig-
nificant, indicating this could be due to the sparse data
for the ”Yes” class. However, to attain this level of
competency as presented in the confusion matrix, a
radiologist would require 3-5 years of experience and
exposure to 2000-5000 patients and 15000-20000
images.
4.2.1 Concordance Score
The concordance score indicates a moderate level
of agreement between the predicted values and the
ground truth. This is consistent with inter-operator
variability among two radiologists. The concordance
score of the ResNet1 model is 0.5835, which reflects
that the model performs reasonably well in predicting
the target variable. The performance can further be
improved by using various data augmentation meth-
ods for a balanced positive class.
5 DISCUSSION
Traditional risk stratification models have often re-
lied on rule-based systems and expert judgment, us-
ing specific clinical factors or markers to estimate the
likelihood of outcomes such as disease progression,
complications, or mortality. These models have been
foundational in clinical settings, helping to prioritize
patient care, allocate resources, and guide testing and
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1090

Figure 7: Confusion Matrix of CNN validation dataset for
csPCa and non-csPCa.
treatment decisions. However, with the emergence of
ML, more dynamic, data-driven approaches have be-
come available, offering the potential for greater ac-
curacy and adaptability in risk assessment.
To develop a reliable and clinically useful model
for patient risk stratification, we employed an image
triage strategy that mixed images from both positive
and negative classes spanning diverse population de-
mographics with respect to age. Results from this
study demonstrate that image-trained models using
MRI images help to triage patients with clinically sig-
nificant prostate cancer with 60% accuracy, highlight-
ing the clinical value and effectiveness of CNNs in
supervised learning. Additionally, machine learning
models trained on patient data yield an accuracy of
69.79%, with both models drawing on information
from the same patients but differing in data types.
The image triage approach enabled the model to learn
more generalizable patterns, capturing subtle varia-
tions and complex features, which enhanced overall
robustness and is a step towards building responsible
and trustworthy AI model.
The confusion matrix results suggest that, despite
applying SMOTE to address the class imbalance in
the dataset, the model’s performance may still be af-
fected. The low TP rate and high FP rate indicate
difficulties in detecting csPCa and a tendency to mis-
classify non-csPCa cases as csPCa. This suggests that
the imbalance, though mitigated by SMOTE, may still
influence the model’s bias toward the majority class
(non-csPCa), and further techniques, such as class
weighting or different resampling methods, may be
needed to improve csPCa detection and reduce false
alarms. By including slices from multiple patients
in both classes, we minimized the risk of bias to-
ward individual-specific characteristics, allowing the
model to reflect patterns representative of the broader
population. Additionally, this strategy helped to mit-
igate the underrepresentation of clinically significant
cancer cases, balancing the training data more effec-
tively. Finally, by incorporating images from different
angles, the model could focus on learning core diag-
nostic features rather than being influenced by slice
positions from different MRI sequences.
5.1 Future Directions
Multi-modal ML approaches, which combine clini-
cal, imaging, and biomarker data, show great promise
in enhancing diagnostic accuracy and patient out-
comes. Studies have demonstrated that integrating
clinical data, serum biomarkers, and imaging tech-
niques can significantly improve diagnostic perfor-
mance, as seen with models like the Irish Prostate
Cancer Risk Calculator (IPRC) and other multi-
platform integrations (Jalali et al., 2020; Mottet et al.,
2021).
Despite these advances, multi-modal ML ap-
proaches for PCa remain underexplored, especially in
the clinical setting. Currently, multi-modal decision-
making occurs primarily in a one-dimensional man-
ner during multidisciplinary team (MDT) meetings,
lacking the integration of ML decision support sys-
tems. This gap highlights an opportunity for fu-
ture research to fully integrate multi-modal data into
ML models, offering a comprehensive and patient-
centered care model. Future directions should focus
on combining diverse data sources—clinical metrics,
imaging data, serum biomarkers, and even patient-
reported outcomes—to refine diagnostic algorithms
and improve the accuracy of distinguishing clinically
significant prostate cancer. Additionally, segmen-
tation techniques and fusion methods could be ex-
plored to enhance the model’s ability to capture in-
tricate patterns across modalities, further boosting di-
agnostic precision. Moreover, the integration of these
models into clinical practice through user-friendly de-
cision support systems will be critical to ensuring
their real-world applicability, improving communica-
tion among healthcare teams, and ultimately leading
to better patient outcomes.
6 CONCLUSION
The study explored two approaches for risk stratifica-
tion of clinically significant prostate cancer without
using biopsy-derived features. The research aimed
to address algorithmic and data-driven biases related
Mitigating Algorithmic Bias in Prostate Cancer Risk Stratification with Responsible Artificial Intelligence and Machine Learning
1091

to patient-specific characteristics. We mitigated data-
driven bias for the age feature and reduced algorith-
mic bias in their image triage approach by mixing
imaging slices from positive and negative classes.
This method allowed the machine learning model
to learn generalizable features, enhancing robustness
and minimizing patient-specific biases. Initial results
suggest the image triage method’s potential to create
more accurate and unbiased classifiers. Future work
will focus on developing a score-based triage system
to assign relevance scores to images based on their in-
formational value for detecting clinically significant
prostate cancer and predicting prostate cancer risk us-
ing behavioral data, such as nutritional features.
REFERENCES
Ageing — oecd.org. https://www.oecd.org/en/topics/
policy-issues/ageing.html. [Accessed 14-11-2024].
Biederer, T. et al. (2016). Simpleitk: A simplified layer
of itk for medical image processing. The Journal of
Digital Imaging, 29(4):32–48.
Brembilla, G., Dell’Oglio, P., Stabile, A., Damascelli, A.,
Brunetti, L., Ravelli, S., Cristel, G., Schiani, E., Ven-
turini, E., Grippaldi, D., et al. (2020). Interreader vari-
ability in prostate mri reporting using prostate imag-
ing reporting and data system version 2.1. European
radiology, 30:3383–3392.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
He, M., Cao, Y., Chi, C., Yang, X., Ramin, R., Wang,
S., Yang, G., Mukhtorov, O., Zhang, L., Kazantsev,
A., et al. (2023). Research progress on deep learn-
ing in magnetic resonance imaging–based diagnosis
and treatment of prostate cancer: a review on the cur-
rent status and perspectives. Frontiers in Oncology,
13:1189370.
Hooshmand, A. (2021). Accurate diagnosis of prostate
cancer using logistic regression. Open Med (Wars),
16(1):459–463.
Jalali, A., Foley, R., Maweni, R., Murphy, K., Lundon,
D., Lynch, T., Power, R., O’Brien, F., O’Malley, K.,
Galvin, D., Durkan, G., Murphy, T., and Watson, R.
(2020). Integrating inflammatory serum biomarkers
into a risk calculator for prostate cancer detection.
BJU International, 125(1):61–68.
Jalali, A., Foley, R., Maweni, R., Murphy, K., Lundon,
D., Lynch, T., Power, R., O’Brien, F., O’Malley, K.,
Galvin, D., Durkan, G., Murphy, T., and Watson, R.
(2023). A risk calculator to inform the need for a
prostate biopsy: a rapid access clinic cohort. Unpub-
lished manuscript.
Jiang, X., Hu, Z., Wang, S., and Zhang, Y. (2023). Dl
for medical image-based cancer diagnosis. Cancers
(Basel), 15(14):3608.
Mottet, N., Bellmunt, J., and Bolla, M. (2017). Eau-estro-
siog guidelines on prostate cancer. European Urology,
71(4):618–629.
Mottet, N. et al. (2021). Eau-eanm-estro-esur-siog guide-
lines on prostate cancer. European Association of
Urology.
Pecoraro, M., Messina, E., Bicchetti, M., Carnicelli, G.,
Del Monte, M., Iorio, B., La Torre, G., Catalano, C.,
and Panebianco, V. (2021). The future direction of
imaging in prostate cancer: Mri with or without con-
trast injection. Andrology, 9(5):1429–1443.
Pellicer-Valero, O. J., Marenco Jimenez, J. L.,
Gonzalez-Perez, V., Casanova Ramon-Borja,
J. L., Mart
´
ın Garc
´
ıa, I., Barrios Benito, M.,
Pelechano Gomez, P., Rubio-Briones, J., Rupe
´
erez,
M. J., and Mart
´
ın-Guerrero, J. D. (2022). Dl for
fully automatic detection, segmentation, and gleason
grade estimation of prostate cancer in multiparametric
magnetic resonance images. Scientific reports,
12(1):2975.
Saha, A., Bosma, J. S., Twilt, J. J., van Ginneken, B.,
Bjartell, A., Padhani, A. R., Bonekamp, D., Villeirs,
G., Salomon, G., Giannarini, G., et al. (2024). Artifi-
cial intelligence and radiologists in prostate cancer de-
tection on mri (pi-cai): an international, paired, non-
inferiority, confirmatory study. The Lancet Oncology.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wo-
jna, Z. (2016). Rethinking the inception architecture
for computer vision. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition,
pages 2818–2826.
Wang, L., Lu, B., He, M., Wang, Y., Wang, Z., and Du,
L. (2022). Prostate cancer incidence and mortality:
Global status and temporal trends in 89 countries from
2000 to 2019. Frontiers in Public Health, 10.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1092
