
Leveraging LLMs and RAG for Schema Alignment:
A Case Study in Healthcare
Rishi Saripalle
1
, Roopa Foulger
2
and Satish Dooda
1
1
School of Information Technology, Illinois State University, Normal, Illinois, U.S.A.
2
Digital and Innovation Development, OSF HealthCare, Peoria, Illinois, U.S.A.
Keywords: Schema Alignment, Schema Integration, Structural Mapping, LLM, Interoperability.
Abstract: In the quest to achieve digital health and enable data-driven healthcare, health organizations often rely on
multiple third-party vendor solutions to monitor and collect patient health and related data, specifically outside
organizations' control, such as home setting, which is later communicated to the organization’s information
systems. However, the reliance on multiple vendor solutions often results in fragmented data structures, as
each vendor solutions system follows its non-standard data model. This fragmentation complicates the data
integration, creating barriers to seamless data exchange and interoperability, which is essential for data-driven
healthcare. Recent advancements in Large Language Models (LLMs) have great potential to analyze data
models and generate rich contextual-semantic metadata for the model, useful for identifying mappings
between disparate data structures. This preliminary research explores the adoption of LLMs in combination
with the Retrieval-Augmented Generation (RAG) approach to facilitate structural alignment between
disparate data models. By semi-automating the schema alignment process—currently a labor-intensive task—
LLMs can streamline the data integration of heterogeneous data models, enhancing efficiency by reducing
the developer’s time and manual effort.
1 INTRODUCTION
As healthcare organizations, specifically hospitals,
continue to digitalize healthcare and adopt data-
driven approaches to enhance clinical and operational
efficiency, improve patient experience, and drive
better health outcomes, the seamless exchange,
integration, and interpretation of data from multiple
sources has become essential. While hospitals make
substantial investments in various information
systems and digital infrastructure, it is neither feasible
nor efficient for them to develop every solution
internally, such as patient monitoring and remote
digital health tools. Consequently, organizations
often rely on multiple third-party vendor solutions to
monitor and collect patient health data, especially in
settings beyond the organization’s immediate control,
such as long-term care facilities and at-home care
environments. These external systems often rely on
proprietary and non-standard data schemas/models to
communicate patient and associated data with the
organization’s information systems. This results in
fragmentation when integrating vendor-captured data
into the hospital's data ecosystem. This fragmentation
poses significant challenges for data integration and
creates barriers to data exchange and interoperability,
which is critical for enabling data-driven activities in
modern healthcare. Harmonizing data across
disparate systems is essential for overcoming these
barriers and ensuring AI and data-driven approaches
can work effectively, allowing the organization to
leverage advanced technologies for improved
decision-making and patient care.
The Data Schema Mapping (DSM) process
identifies correspondences between elements of
different data schemas to facilitate data integration,
interoperability, or migration. It plays a crucial role in
relational databases, information exchange, and
ontologies, mainly as data grows more complex and
interconnected. DSM is essential for structurally and
semantically linking data, enhancing search and
retrieval processes, and enabling seamless integration
between disparate data models. However, manually
mapping schemas across multiple systems is both
intensive and time-consuming, requiring significant
expertise and impractical to scale for large-scale
projects. From an organization’s standpoint, relying
on highly qualified professionals' time for schema
750
Saripalle, R., Foulger, R. and Dooda, S.
Leveraging LLMs and RAG for Schema Alignment: A Case Study in Healthcare.
DOI: 10.5220/0013262800003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 750-757
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

mapping is financially inefficient. Consequently,
with technological advances, researchers have
increasingly turned to Machine Learning (ML) and
Natural Language Processing (NLP) techniques to
automate and improve the DSM process. Using NLP,
xMatcher (Yousfi et al., 2020) identifies semantically
similar schema elements by understanding the
meaning of schema elements, uses WordNet to
associate elements, and computes semantic similarity
between elements. A similar approach was adopted
for integrating energy data (Pan et al., 2022) and
mapping XML schemas (Fan et al., 2016). Linguistics
analysis, language structures, and NLP techniques
combined are used for schema (any format) matching
(Li, 2020; J. Zhang et al., 2021), integrating and
migrating data in the cloud (Chandak et al., 2024),
combining protein crystallization experimental data
from different labs (Shrestha et al., 2020), and
mapping food terms/words to nutrition concepts
(Stojanov et al., 2020). Using AI/ML, relational
database schemas are merged (Sahay et al., 2020; Y.
Zhang et al., 2023; Zhou et al., 2024), and ontology
integration (Adithya et al., 2022; Feng & Fan, 2019).
By leveraging AI/ML and NLP, the DSM process can
now account for linguistic and semantic similarities,
structural patterns, and contextual information across
disparate data schema/model(s), allowing for a more
intelligent and scalable approach.
While ML and NLP techniques have significantly
improved the schema-matching process, they have
certain limitations. ML models require training
datasets and require extensive training or rely on
domain-specific fine-tuning to achieve desired
results, which is resource-intensive and time-
consuming. While effective in processing linguistic
patterns, NLP techniques need help understanding
complex semantics, predefined rules, heuristics, and
linguistic features, limiting their ability to capture
nuanced meanings and contextual relationships
between schema elements/attributes. This
dependence on extensive feature engineering and
domain expertise makes NLP techniques less scalable
and adaptable to diverse data sources. These
limitations hinder ModelOps (Gartner, 2020) —
particularly in managing data and context drift,
further complicating the deployment and
maintenance of these technologies.
Recently, Large Language Models (LLMs),
powered by advanced AI and trained on extensive
datasets, have demonstrated remarkable capabilities
in understanding and inferring the context and
semantics of data while also managing the subtleties
of language with great precision. Unlike ML and NLP
techniques, LLMs grasp semantics relationships and
infer context without requiring large training datasets
or fine-tuning, offering a more flexible and scalable
solution. This raises the question: can LLMs enhance
the DSM process or its tasks by overcoming the
limitations of the earlier approaches?
As an emerging area of research, initial attempts
have been made to use LLMs for DSM (Kiourtis et
al., 2019; Satti et al., 2021; Sett et al., 2024; Sheetrit
et al., 2024). These studies leveraged LLMs language
processing and contextual understanding capabilities
along with the schema (e.g., description, context of
use) and their elements metadata (e.g., description,
data type, relationships with other elements) to derive
mappings between schemas. In these studies, schema
metadata, particularly the element descriptions, are
transformed into embeddings using embedding
models and stored in a vector database. This enables
a semantic search, where attributes from a source
schema can be queried to find semantically similar
target attributes by retrieving documents that meet a
specific similarity threshold. The success of this
process heavily depends on the availability and
quality of attribute descriptions, which serve as the
primary input for generating accurate and relevant
embeddings. These studies predominantly relied on
the availability and quality of descriptions to drive the
mapping process.
While these early studies show promise,
highlighting the potential of LLMs to enhance the
DSM process, they exhibit notable drawbacks. First,
they predominantly rely on the availability and
quality of descriptions to drive the mapping process,
which can be problematic in real-world scenarios
where metadata is sparse, incomplete, or poorly
defined. Second, previous attempts didn’t leverage
Retrieval-Augmented Generation (RAG) and prompt
engineering to provide improved context when
utilizing LLMs. Finally, none of these studies tested
their solutions on real-world schemas commonly
encountered in the hospital data ecosystems. These
drawbacks underscore the need for a methodology to
effectively navigate these challenges and strengthen
the adaptability of the DSM process for real-world
settings.
This paper investigates the potential of LLMs
with the RAG for the DSM. By integrating LLMs
with the RAG and using prompt engineering (Google,
2024; J. Wang et al., 2024; White et al., 2023), we
aim to establish a DSM process that reduces the
manual effort placed on skilled professionals. The
paper is organized as follows. Section 2
(Methodology) outlines the proposed approach for
the DSM using LLMs and RAG. Section 3 (Results
and Evaluation) presents the results obtained from
Leveraging LLMs and RAG for Schema Alignment: A Case Study in Healthcare
751
applying our approach to know schemas and their
mapping in healthcare and real-world scenarios to
validate effectiveness. Finally, Section 4 (Discussion)
and Section 5 (Conclusion) examine our findings,
discuss the limitations, and provide concluding
insights into future work.
2 METHODOLOGY
While previous research studies have relied on the
availability of rich metadata for schemas and their
elements, real-world scenarios are often different and
present significant challenges. Vendor solutions may
not always provide detailed documentation or
comprehensive metadata for the data schema(s) used
in their responses. Instead, they typically offer only a
set of sample data responses corresponding to various
data requests. These samples serve mainly as
developer aids for understanding data structure but
lack the critical metadata and context necessary for
deeper interpretation. This absence of contextual
knowledge makes it difficult to deduce the purpose
and meaning of data fields for secondary uses, such
as Data Schema Matching (DSM), where accurate
field alignment across different schemas is essential.
To address this limitation, our approach focuses
on either generating suitable metadata for the schema
elements or enhancing the existing metadata. This
enhancement aims to improve the accuracy of the
DSM process, ensuring the derived mappings
between schemas are more reliable and reflective of
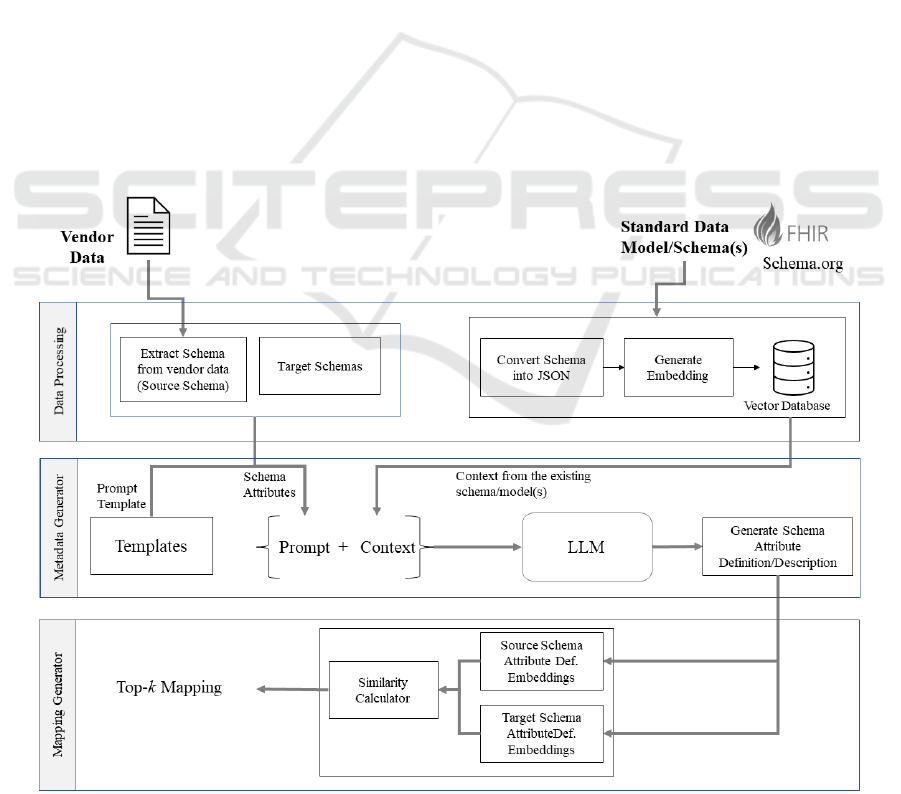
real-world applications. Figure 1 renders our
methodology, which has three stages: Data
Processing, Metadata Generator, and Mapping
Generator.
2.1 Data Processing
The initial step in the proposed DSM approach,
similar to many AI/ML approaches, begins with data
processing. To begin, in case the vendor does not
provide a schema, the vendor-provided response data
is used to derive a schema to map vendor data with
the organization's data ecosystem. Both the source
schema (from the response data) and the target
schema (from the organization's data ecosystem) are
processed and represented in JSON format. In this
JSON structure, a key represents a schema element's
name and has an associated value that is described by
three primary attributes: parent, holds the name of the
parent key, if any; default, holds original metadata
Figure 1: Overall architecture of Data Schema Mapping processing using LLM with RAG.
HEALTHINF 2025 - 18th International Conference on Health Informatics
752
provided by the vendor or the source schema; and rag,
holds the metadata generated by our approach. For
nested elements, the key name is constructed by
concatenating the parent element's name with the
element's name, ensuring each nested key remains
unique. The presented approach uses RAG to provide
context to the LLM for generating better and accurate
metadata for the schema elements. To this end, the
proposed approach has utilized FHIR Resources
(International (HL7), 2019) and schemas from
Schema.org. Schema.org (Guha et al., 2016;
Schema.org, 2024) provides a standard set of schemas
for common vocabulary for entities (e.g., Drug,
Article, Person, Event, MedicalCondition, etc.) and
actions (e.g., Action, MoveAction, etc.) designed to
enable interoperability across the web and improve
search optimization. These schemas allow consistent
data structure, supporting easier data sharing and
integration across various domains. By leveraging
Schema.org's and HL7 FHIR’s well-defined metadata
on schema/resources and its elements, the proposed
approach ensures that the context fed into the LLM is
comprehensive and consistent. The approach
leverages the LangChain framework (LangChain,
2023), Schema.org, and the HL7 FHIR standard to
implement the RAG. The Schema.org schemas and
HL7 FHIR (R4) resources are translated into separate
JSON documents. One JSON document is dedicated
to FHIR resources, where each key represents either
a resource name (e.g., Patient, Observation, etc.) or a
resource attribute (e.g., name, contact, etc.), and its
corresponding value is the description of that resource
or resource attribute. Similarly, Schema.org entities
and actions are organized into another JSON
document following the same structure. Here, each
key corresponds to an entity or action name or an
attribute of an entity/action, with the value being the
respective description. Using the LangChain
document loader, the two JSON documents are
recursively divided (using RecursiveJsonSplitter)
into manageable chunks (chunk size of 300) and
loaded in a FAISS vector database using the OpenAI
embedding model (OpenAI, 2022) – “text-
embedding-3-small” is used in this approach. This
enables efficient storage (used in memory storage)
and retrieval (used Maximal Marginal Relevance
strategy and search threshold of .7) of contextual
knowledge, facilitating the RAG approach and
providing relevant context to the LLM to generate
accurate descriptions of schema elements.
2.2 Metadata Generator
The goal of this stage in the DSM process, Figure 1,
is to generate metadata, specifically, the description
of the schema elements, using the structure of the
schema, the contextual knowledge from standard
schemas (Schema.org and FHIR), and LLM (GPT-4)
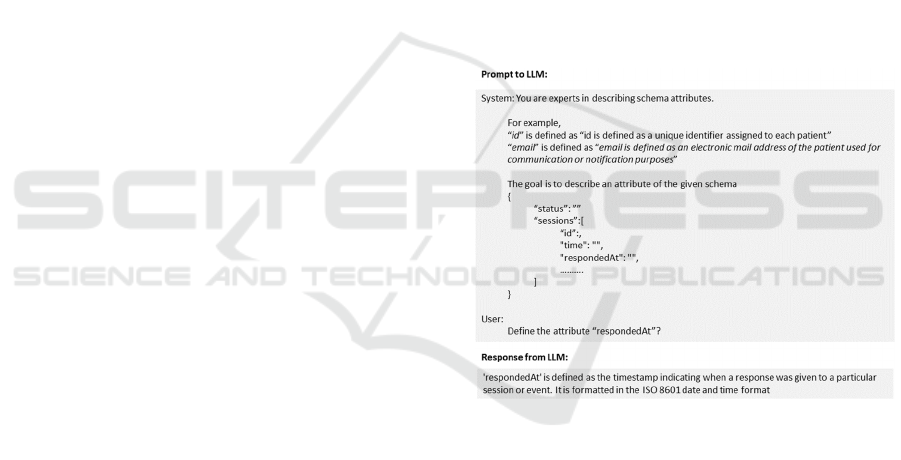
itself. The prompt requesting the LLM to describe a
schema element must be carefully structured and
well-formulated to obtain an accurate response, a
process known as prompt engineering. Following
best practices and guidelines for prompt engineering
(Google, 2024), the proposed approach uses the Few-
Shot prompting technique and RACE (Role-Action-
Expectation) prompt structure to write the prompt to
elicit high-quality descriptions of schema elements,
thus improving the overall efficiency and
effectiveness of the methodology. The generated
schema element descriptions are then incorporated
into the JSON document, created during the Data
Processing, and associated with the schema,
enriching the metadata. Figure 2 illustrates an
example of a prompt and the corresponding LLM
response.
Figure 2: Prompt to LLM to generate metadata for a schema
element using Prompt Engineering and RACE prompt
structure.
2.3 Mapping Generator
The final stage of the DSM process involves aligning
the source and target schema elements. Using the
descriptions generated in the previous stage,
embedding for all the source and target schema
elements is generated using the "all-MiniLM-L6-v2"
(Transformers, 2020; W. Wang et al., 2020) Sentence
transformer model. This model is used as it efficiently
produces dense embeddings for short texts, allowing
for an accurate semantic comparison. By calculating
the similarity, cosine similarity, between the
embeddings, this stage effectively identifies and
aligns schema elements with the highest contextual
Leveraging LLMs and RAG for Schema Alignment: A Case Study in Healthcare
753

and semantic relevance, enhancing the precision of
the mappings of the DSM process. The following is
the algorithm that generates MappingK elements
from the target schema that are semantically similar
to each attribute in the source schema:
Algorithm 1: Caption example.
Data: Source Schema Elements (S
S
); Target Schema
Elements (T
S
); Embedding model Φ; M is the
size of S
E
and N is the size of T
E
where M, N
∈ N
Results: M x N cosine similarity matrix (T
CS
); M x K
matrix (T
K
) with Mapping-K elements with
highest similarity between S
S
and T
S
foreach s
i
∈ S
E
do
s
ie =
Φ (s
i
)
foreach t
j
∈ T
E
do
t
je =
Φ (s
j
)
T
CS
cosineSimilarity (s
ie,
t
je
)
end
end
T
K
CreateMappingK(T
CS
,K)
3 EVALUATION
We used two mapping datasets to evaluate the
proposed approach: MIMIC-III to OMOP (Sheetrit et
al., 2024) and Synthea to OMOP (J. Zhang et al.,
2021), providing a standardized benchmark for the
proposed approach. Two OSF hospital information
ecosystem schemas were used to test the approach for
real-world scenarios. Table 1 shows the statistics of
the standardized datasets. The MIMIC, Synthea, and
OMOP standards models are structured as tables and
columns, and their mappings are well-documented
(https://ohdsi.github.io/Tutorial-ETL/ and https://
github.com/meniData1/MIMIC_2_OMOP)
Table 1: MIMIC and Synthea models. Columns and Tables
refer to the number of columns and tables in the models.
Mappings refer to the total number of unique mappings
(removed foreign keys) for the source to target (OMOP).
None refers to columns without mappings between source
and target.
Dataset Columns Tables Mappings None
MIMIC 299 26 - -
MIMC
(OMOP)
- - 89 112
Synthea 155 13 - -
Synthea
(OMOP)
- - 51 116
As explained in Section 2.3, our approach
generates text embedding of the schema elements
metadata and leverages cosine similarity — yielding
a value between 0 and 1— to assess the semantic
similarity between two embeddings. A value closer to
1 indicates higher similarity between the metadata,
suggesting a stronger alignment between the schema
elements, while a lower value signifies reduced
similarity, suggesting a weaker alignment.
Furthermore, this study doesn't consider many-to-
many matches and only looks for one-on-one
mappings between source and target elements. The
simple metric used for evaluation is mapping@k,
similar to a previous study (Sheetrit et al., 2024).
mapping@1 denotes a positively identified mapping
between source and target with the highest similarity
score, mapping@2 denotes a positively identified
mapping with the second highest similarity score,
implying the mapping at one is not accurate or
appropriate, and so forth. A lower K value indicates a
stronger alignment between source and target
elements, with mapping@1 being the ideal outcome.
For each element from the source schema, our process
ranks potential target schema elements based on
similarity score, facilitating automatic alignment
between the schemas.
The MIMIC, Synthea, and OMOP standard
models are thoroughly documented, providing
detailed descriptions of their tables and columns. To
have a baseline for comparison, we have used the
standards’ provided default metadata to generate the
mapping between source (MIMC/Synthea) and
target (OMOP) using only the Mapping Generator
process, which involves generating the embedding
for the metadata and calculating semantic similarity
score. Later, we used our RAG approach, starting
with the Data Processing stage, to generate the
metadata for the columns. Table 2 shows the
mapping@5 results of mapping between MIMIC
and OMOP models. Using the default metadata from
MIMIC and OMOP, we identified 21 mappings,
while our RAG approach yielded 28 mappings with
the highest semantic similarity score. Table 3 shows
the mapping@5 between Synthea and OMOP,
showing 17 mappings that were identified using
default metadata and 19 mappings with the RAG
approach, which had the highest semantic similarity
score.
HEALTHINF 2025 - 18th International Conference on Health Informatics
754

Table 2: mapping@5 mapping between MIMC and OMOP.
%C represents the cumulative percentage of the number of
mappings identified within mapping@5 relative to all
identified mappings.
@1 @2 @3 @4 @5 %C
Default
metadata
21 13 2 10 4 56.17
Using
RAG
28 30 8 4 4 71.9
Table 3: mapping@5 mapping between Synthea and
OMOP. %C represents the cumulative percentage of the
number of mappings identified within mapping@5 relative
to all identified mappings.
@1 @2 @3 @4 @5 %C
Default
metadata
17 6 3 0 1 82.35
Using
RAG
19 3 1 3 1 82.35
The results indicate that metadata generated using
our RAG approach notably improved mapping
accuracy, especially within the top two ranks.
Approximately 50% of the mappings were identified
in the mapping@2, suggesting that supplying the
LLM with richer, contextually relevant metadata
through the RAG approach enhances its
understanding of schema elements. This
improvement contributes to identifying accurate
mappings between schema elements, demonstrating
the potential of enriched metadata in improving
schema alignment accuracy. We have tested the
approach on two CareSignal (CareSignal,
Lightbeam’s Deviceless Remote Patient Monitoring®
Solution, n.d.) schemas—extracted from the API
response data for this research— each mapped to
corresponding OSF schemas. The metadata for the
schemas and their elements was generated using our
approach, as the existing metadata is incomplete and
not suitable for semantic analysis and data integration
purposes. All the schemas are represented in JSON
format (section 2.1), and the number of unique
mappings between the schemas and the proposed
methodology-generated mappings are verified
manually. The vendor schema Schema_V1 containing
61 elements is mapped to OSF_V1 schema with 52
elements. These two schemas have 15 unique
mappings. Similarly, Schema_V2 with 62 elements is
mapped to OSF V2 with 52 elements with 52 unique
mappings. Table 4 shows the results. It should be
noted that the higher mapping accuracy may stem
from the meaningful label names and similar element
names across the source (CareSignal API) and target
(OSF) schemas. However, it is essential to emphasize
that the mappings are identified based on the semantic
similarity of the descriptions generated using RAG
rather than any linguistic or String similarity
techniques between the element names.
Table 4: mapping@5 mapping between CareSignal
schemas and OSF schemas. %C represents the cumulative
percentage of the number of mappings identified within
mapping@5 relative to all identified mappings.
@1 @2 @3 @4 @5 %C
Schema V1 12 0 0 0 0 80
Schema V2 45 0 1 1 0 90.38
4 DISCUSSION
Schema matching is a complex task, complicated by
the heterogeneity across schemas, semantic
ambiguity of schema elements, lack of proper
documentation, and the intricate structure of large
information systems. This challenge is further
compounded by the need for domain-specific
knowledge and limited contextual information for
interpreting the schema elements. Our approach using
LLM with RAG shows a promising preliminary step
towards leveraging LLMs to understand schema
structure, generate appropriate schema element
metadata, and align schemas. The proposed approach
achieves high accuracy in practice without requiring
labeled data, extensive schema-matching datasets, or
the actual data — relying only on the metadata —
highlighting its potential for real-world applications.
For data engineers, this approach reduces the time and
effort for DSM, specifically when fifty percent of the
mappings are identified at mapping@2. Overall, this
enables faster and more efficient schema alignment
and data integration, allowing data engineers to focus
on higher-value tasks rather than manual mapping
efforts.
This preliminary study has notable limitations.
The effectiveness of our approach is closely tied to
the meaningful labels of the schema elements. While
the LLM successfully generated metadata, its
effectiveness depends on schema elements having
meaningful labels recognizable by the LLM, allowing
it to generate accurate and meaningful descriptions
later used for embedding and similarity measure. For
instance, a few element names in the vendor schema
were custom acronyms specific to the response data,
resulting in metadata generation that was either
overly generic or inaccurate, impacting the similarity
score. This observation is broadly applicable to
schemas in other domain areas as well, where the use
of domain-specific acronyms or shorthand often
poses challenges for metadata generation and schema
Leveraging LLMs and RAG for Schema Alignment: A Case Study in Healthcare
755

matching. Hence, when element names lack clarity or
contain acronyms or shorthand terms, the quality of
the generated metadata and the accuracy of schema
matching is compromised. Furthermore, the
mapping@K metric needs to be adapted or replaced
with better evaluation metrics for one-to-many and
many-to-many mappings. Nonetheless, this study
lays the groundwork for further exploring the use of
LLMs and their advanced language capabilities in
data schema mapping, enabling interoperability
across data systems.
5 CONCLUSIONS
This project demonstrates an approach to the DSM
process by utilizing LLMs with RAG to semi-
automate schema alignment. By tapping into the
advanced language capabilities of LLMs to generate
metadata and using text embedding models to identify
semantically similar elements, the approach reduces
the manual effort required for schema alignment.
Notably, this method does not require predefined
mappings, model training, or direct access to source
data, making it highly adaptable. Although it faces
limitations related to metadata quality and
dependency on meaningful schema element labels,
this preliminary study sets a strong foundation for
further exploration of the LLM-based DSM process.
For future work, the avenues for advancement
include extending the current approach to address
many-to-many mappings and exploring domain-
specific fine-tuning of LLMs, which could enhance
the process’s ability to interpret and generate accurate
metadata for domain-specific terminology,
specifically acronyms. Additionally, integrating
human feedback mechanisms into the DSM process
could iteratively refine the LLM's contextual
understanding, leading to progressively improved
accuracy and effectiveness in schema matching over
time.
ACKNOWLEDGMENTS
This research was made possible with funding from
the Connected Communities Initiative (CCI), a
collaborative initiative between Illinois State
University and OSF HealthCare. The authors also
wish to acknowledge Safura Sultana for project
management and Navya Godavarthi, Kevin
Gustafson, Richard Hall, and David McGrew for
valuable feedback and data assistance. This project
was approved as non-human subjects research by the
Peoria IRB.
REFERENCES
Adithya, V., Deepak, G., & Santhanavijayan, A. (2022).
OntoIntAIC: An Approach for Ontology Integration
Using Artificially Intelligent Cloud. In P. Verma, C.
Charan, X. Fernando, & S. Ganesan (Eds.), Advances
in Data Computing, Communication and Security (Vol.
106, pp. 3–13). Springer Nature Singapore.
https://doi.org/10.1007/978-981-16-8403-6_1
CareSignal, Lightbeam’s Deviceless Remote Patient
Monitoring® solution. (n.d.). [Computer software].
Lightbeam Health Solutions. https://lightbeamhealth.
com/deviceless-remote-patient-monitoring
Chandak, M., Rathi, S., Chordiya, H., Rawat, S.,
Rachapotu, S. V. K., & Barodia, U. (2024). An NLP-
Based Approach for Data Integration and Migration on
Cloud Infrastructure. In L. Garg, N. Kesswani, I.
Brigui, B. Kr. Dewangan, R. N. Shukla, & D. S. Sisodia
(Eds.), AI Technologies for Information Systems and
Management Science (Vol. 1071, pp. 1–12). Springer
Nature Switzerland. https://doi.org/10.1007/978-3-
031-66410-6_1
Fan, H., Deng, K., & Liu, J. (2016). An Approach of XML
Schema Matching Using Top-K Mapping. 2016 3rd
International Conference on Information Science and
Control Engineering (ICISCE), 174–178. https://doi.
org/10.1109/ICISCE.2016.47
Feng, Y., & Fan, L. (2019). Ontology semantic integration
based on convolutional neural network. Neural
Computing and Applications, 31(12), 8253–8266.
https://doi.org/10.1007/s00521-019-04043-w
Gartner. (2020). ModelOps. Gartner. https://www.gartner.
com/en/information-technology/glossary/modelops
Google. (2024). What is Prompt Engineering?
https://cloud.google.com/discover/what-is-prompt-
engineering
Guha, R. V., Brickley, D., & Macbeth, S. (2016).
Schema.org: Evolution of structured data on the web.
Communications of the ACM, 59(2), 44–51.
https://doi.org/10.1145/2844544
International (HL7), H. L. S. (2019). FHIR Release 4 (R4).
Health Level Seven International. https://hl7.org/FHIR/
Kiourtis, A., Mavrogiorgou, A., Menychtas, A.,
Maglogiannis, I., & Kyriazis, D. (2019). Structurally
Mapping Healthcare Data to HL7 FHIR through
Ontology Alignment. Journal of Medical Systems,
43(3), 62. https://doi.org/10.1007/s10916-019-1183-y
LangChain. (2023). LangChain: Building Applications with
LLMs through Composability. LangChain.
https://www.langchain.com/
Li, G. (2020). DeepFCA: Matching Biomedical Ontologies
Using Formal Concept Analysis Embedding
Techniques. Proceedings of the 4th International
Conference on Medical and Health Informatics, 259–
265. https://doi.org/10.1145/3418094.3418121
HEALTHINF 2025 - 18th International Conference on Health Informatics
756

OpenAI. (2022). Text-embedding-ada-002. OpenAI.
https://platform.openai.com/docs/guides/embeddings
Pan, Z., Pan, G., & Monti, A. (2022). Semantic-Similarity-
Based Schema Matching for Management of Building
Energy Data. Energies, 15(23), 8894. https://doi.
org/10.3390/en15238894
Sahay, T., Mehta, A., & Jadon, S. (2020). Schema Matching
using Machine Learning. 2020 7th International
Conference on Signal Processing and Integrated
Networks (SPIN), 359–366. https://doi.org/10.
1109/SPIN48934.2020.9071272
Satti, F. A., Hussain, M., Hussain, J., Ali, S. I., Ali, T., Bilal,
H. S. M., Chung, T., & Lee, S. (2021). Unsupervised
Semantic Mapping for Healthcare Data Storage
Schema. IEEE Access, 9, 107267–107278.
https://doi.org/10.1109/ACCESS.2021.3100686
Schema.org. (2024). Schema vocabulary for structured
data on the Internet (Version 28) [Computer software].
Schema.org. https://schema.org/
Sett, A., Hashemifar, S., Yadav, M., Pandit, Y., & Hejrati,
M. (2024). Speaking the Same Language: Leveraging
LLMs in Standardizing Clinical Data for AI (No.
arXiv:2408.11861). arXiv. http://arxiv.org/abs/2408.
11861
Sheetrit, E., Brief, M., Mishaeli, M., & Elisha, O. (2024).
ReMatch: Retrieval Enhanced Schema Matching with
LLMs (No. arXiv:2403.01567). arXiv. http://arxiv.
org/abs/2403.01567
Shrestha, M., Tran, T. X., Bhattarai, B., Pusey, M. L., &
Aygun, R. S. (2020). Schema Matching and Data
Integration with Consistent Naming on Protein
Crystallization Screens. IEEE/ACM Transactions on
Computational Biology and Bioinformatics, 17(6),
2074–2085. https://doi.org/10.1109/TCBB.2019.
2913368
Stojanov, R., Kocev, I., Gramatikov, S., Popovski, G.,
Korousic Seljak, B., & Eftimov, T. (2020). Toward
Robust Food Ontology Mapping. 2020 IEEE
International Conference on Big Data (Big Data),
3596–3601. https://doi.org/10.1109/BigData50022.
2020.9378066
Transformers, S. (2020). All-MiniLM-L6-v2 [Computer
software]. https://huggingface.co/sentence-transformers/
all-MiniLM-L6-v2
Wang, J., Shi, E., Yu, S., Wu, Z., Ma, C., Dai, H., Yang, Q.,
Kang, Y., Wu, J., Hu, H., Yue, C., Zhang, H., Liu, Y.,
Pan, Y., Liu, Z., Sun, L., Li, X., Ge, B., Jiang, X., …
Zhang, S. (2024). Prompt Engineering for Healthcare:
Methodologies and Applications (No.
arXiv:2304.14670). arXiv. http://arxiv.org/abs/2304.
14670
Wang, W., Wei, F., Dong, L., Bao, H., Yang, N., & Zhou,
M. (2020). MiniLM: Deep Self-Attention Distillation
for Task-Agnostic Compression of Pre-Trained
Transformers (No. arXiv:2002.10957). arXiv.
http://arxiv.org/abs/2002.10957
White, J., Fu, Q., Hays, S., Sandborn, M., Olea, C., Gilbert,
H., Elnashar, A., Spencer-Smith, J., & Schmidt, D. C.
(2023). A Prompt Pattern Catalog to Enhance Prompt
Engineering with ChatGPT (No. arXiv:2302.11382).
arXiv. http://arxiv.org/abs/2302.11382
Yousfi, A., Hafid, M., & Zellou, A. (2020). xMatcher:
Matching Extensible Markup Language Schemas using
Semantic-based Techniques. International Journal of
Advanced Computer Science and Applications,
11(8).
https://doi.org/10.14569/IJACSA.2020.0110880
Zhang, J., Shin, B., Choi, J. D., & Ho, J. C. (2021). SMAT:
An Attention-Based Deep Learning Solution to the
Automation of Schema Matching. In L. Bellatreche, M.
Dumas, P. Karras, & R. Matulevičius (Eds.), Advances
in Databases and Information Systems (Vol. 12843, pp.
260–274). Springer International Publishing. https://
doi.org/10.1007/978-3-030-82472-3_19
Zhang, Y., Floratou, A., Cahoon, J., Krishnan, S., Müller,
A. C., Banda, D., Psallidas, F., & Patel, J. M. (2023,
January). Schema Matching using Pre-Trained
Language Models. ICDE. https://www.microsoft.
com/en-us/research/publication/schema-matching-using
-pre-trained-language-models/
Zhou, X., Dhingra, L. S., Aminorroaya, A., Adejumo, P., &
Khera, R. (2024). A Novel Sentence Transformer-based
Natural Language Processing Approach for Schema
Mapping of Electronic Health Records to the OMOP
Common Data Model. Health Informatics.
https://doi.org/10.1101/2024.03.21.24304616.
Leveraging LLMs and RAG for Schema Alignment: A Case Study in Healthcare
757
