
Preliminary Results on Using Clustering of Functional Data to Identify
Patients with Alzheimer’s Disease by Analyzing Brain MRI Scans
Calin Anton
1
, Cristina Anton
2
, Mohamad El-Hajj
1
, Matthew Craner
1
and Richard Lui
1
1
Department of Computer Science, MacEwan University, Edmonton, Alberta, Canada
2
Department of of Mathematics and Statistics, MacEwan University, Edmonton, Alberta, Canada
{antonc, popescuc, elhajjm}@macewan.ca, {cranerm2, luir}@mymacewan.ca
Keywords:
Clustering of Functional Data, Brain MRI, Alzheimer’s Disease.
Abstract:
This study delves into the effectiveness of funWeightClust, a sophisticated model-based clustering technique
that leverages functional linear regression models to pinpoint patients diagnosed with Alzheimer’s Disease.
Our research entailed a thorough analysis of voxelwise fractional anisotropy data derived from the Alzheimer’s
Disease Neuroimaging Initiative (ADNI) dataset, with a particular emphasis on the Cingulum and Corpus Cal-
losum, which are critical regions of interest in understanding the disease’s impact on brain structure. Through
a series of experiments, we established that funWeightClust is efficient at distinguishing between patients
with Alzheimer’s Disease and healthy control subjects. Notably, the clustering model yielded even more
pronounced and accurate results when we focused our analysis on specific brain regions, such as the Left
Hippocampus and the Splenium. We postulate that integrating additional biomarkers could significantly en-
hance the accuracy and reliability of funWeightClust in identifying patients who exhibit signs of Alzheimer’s
Disease.
1 INTRODUCTION
Alzheimer’s Disease (AD) is a complex and chronic
neurodegenerative disorder that primarily impacts the
brain, leading to a progressive decline in cognitive
functions such as memory, reasoning, and overall be-
havior. Unlike the natural aging process, Alzheimer’s
is not a typical consequence of aging and is character-
ized by its irreversible nature, meaning that once the
disease sets in, it cannot be reversed or cured.
This condition is the most common form of de-
mentia, accounting for an estimated 60% to 80% of all
dementia cases worldwide. The disease is marked by
the accumulation of amyloid plaques and tau tangles
in the brain, which ultimately disrupt communication
between neurons and result in cell death.
According to a comprehensive report by the
Alzheimer Society of Canada (Armstrong et al.,
2022), the prevalence of dementia within the Cana-
dian population is anticipated to escalate significantly
in the coming years. It is projected that nearly 1 mil-
lion individuals in Canada could be living with de-
mentia by the year 2030, leading to an alarming in-
crease of approximately 187,000 new cases annually.
Furthermore, by the year 2050, estimates suggest that
this number could rise to more than 1.7 million Cana-
dians affected by dementia, highlighting the urgent
need for enhanced awareness, research, and resources
to address this growing health crisis.
Grasping the progression of Alzheimer’s disease
is essential for facilitating early detection, implement-
ing effective treatment strategies, and ultimately en-
hancing the quality of life for those impacted by the
condition, as well as their families. The diagnosis of
AD involves a comprehensive approach that includes
detailed clinical evaluations, the identification of spe-
cific biomarkers, advanced brain imaging techniques,
and thorough neuropsychological assessments. Sig-
nificant advancements in any of these domains can
greatly improve the efficiency and accuracy of early
AD detection, paving the way for timely interventions
that can make a substantial difference in patient out-
comes.
Numerous papers have explored the use of ma-
chine learning and deep learning techniques for diag-
nosing Alzheimer’s Disease (Dara et al., 2023). These
studies cover a range of topics, including compar-
isons between cognitively normal (CN) individuals
and those diagnosed with AD, as well as comparisons
between CN subjects and those with mild cognitive
impairment (MCI). As a result, the accuracy of find-
ings across these studies varies significantly, ranging
Anton, C., Anton, C., El-Hajj, M., Craner, M. and Lui, R.
Preliminary Results on Using Clustering of Functional Data to Identify Patients with Alzheimer’s Disease by Analyzing Brain MRI Scans.
DOI: 10.5220/0013263500003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 363-368
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
363

from as low as 30% to as high as 90%.
The survey by Kaur et al (Kaur et al., 2024) inves-
tigates various techniques for detecting Alzheimer’s
disease, focusing on datasets, input modalities, algo-
rithms, libraries, and performance metrics to identify
the most effective strategies. The study analyzed 100
research articles published between 2019 and 2022. It
found that most studies used deep learning strategies,
with datasets primarily from the Alzheimer’s Disease
Neuroimaging Initiative (ADNI). Convolutional Neu-
ral Networks achieved the highest accuracy (100%)
in classifying AD vs. CN subjects, while Support
Vector Machines were the most frequently used ma-
chine learning algorithm, with a maximum accuracy
of 99.82%.
Diffusion Tensor Imaging (DTI) is regarded as
one of the most effective methods for detecting AD
in patients (Oishi et al., 2011). A study examining
DTI indicators of white matter impairment associated
with AD was conducted in (Nir et al., 2013). The
study concluded that AD patients exhibited clear dis-
ruptions in anisotropy and diffusivity, particularly in
the cingulum and corpus callosum.
The study conducted in (Schouten et al., 2017) ex-
plored the use of fractional anisotropy (FA) indepen-
dent components analysis (ICA) as a potential pro-
tocol for classifying Alzheimer’s disease. In this re-
search, FA, mean diffusivity, axial diffusivity, and ra-
dial diffusivity were independently utilized for clus-
tering through independent components, aiming to
extract the mixing weights. The classification method
presented achieved an accuracy of 85%. The authors
also noted that determining the required weightings
for ICA can be challenging, particularly in extended
studies.
In (Ma et al., 2019), researchers investigate the
relationship between Mini-Mental State Examination
(MMSE) scores and brain imaging data from various
regions of interest (ROIs), along with multiple single
nucleotide polymorphisms across different quantiles.
The findings indicate that the left thalamus, left hip-
pocampus, and right lateral ventricle are the most sig-
nificant ROIs. Additionally, the study highlights that
education level and age are the key factors influencing
MMSE scores.
Clusterwise functional linear regression models
are employed in (Li et al., 2021) to explore the re-
lationship between fractional anisotropy curves along
the cingulum and the body of corpus callosum skele-
tons with Mini-Mental State Examination scores in
Alzheimer’s disease and cognitively normal patients
from the ADNI (adni.loni.usc.edu) dataset. The most
effective methods achieve a Rand index of 0.9 and an
adjusted Rand index of 0.8.
In this paper, we explore the effectiveness of fun-
WeightClust. This innovative model-based clustering
technique leverages a mixture of functional linear re-
gression models specifically designed for identifying
subjects with Alzheimer’s disease. One of the key
strengths of funWeightClust is its capability to man-
age complex functional multivariate responses and
predictors, making it well-suited for the nuanced na-
ture of medical data. To evaluate the performance of
funWeightClust, we utilize a comprehensive dataset
obtained from ADNI. Our preliminary findings sug-
gest that this novel method significantly enhances the
ability to differentiate between individuals diagnosed
with AD and those in the control group, indicating its
potential as a valuable tool in clinical diagnostics.
2 METHODS
2.1 Data Collection and Processing
We conducted a detailed analysis of the functional
data derived from pre-processed Diffusion Tensor
Imaging scans from the Alzheimer’s Disease Neu-
roimaging Initiative data set. Our research focused
on two distinct groups: individuals diagnosed with
Alzheimer’s disease and cognitively normal partic-
ipants. The AD group comprised 75 participants
whose ages ranged from 62 to 92 years. Within this
group, there were 50 males and 25 females.
In contrast, the CN group included 137 partici-
pants, with ages spanning from 60 to 93 years. This
group contained 74 males and 63 females, providing
a balanced representation of both genders.
This comprehensive data collection allows for
a nuanced exploration of the differences in brain
structure and function between individuals with
Alzheimer’s disease and cognitively healthy individ-
uals.
Data imbalance can adversely impact the perfor-
mance of functional clustering algorithms. To address
the class imbalance, we use the adjusted Rand index
(ARI) as a measure of clustering accuracy. This en-
sures that our clustering results are reliable despite the
inherent data imbalances.
We selected the corrected FA images for each
subject and applied skeletonization using the TBSS-
ENIGMA pipeline. The TBSS-ENIGMA function
is part of the ENIGMA (Enhancing NeuroImaging
Genetics through Meta-Analysis) DTI project, which
aims to standardize and improve the analysis of dif-
fusion tensor imaging data across multiple sites (Ja-
hanshad et al., 2013). TBSS (Tract-Based Spatial
Statistics) (Smith et al., 2006) is a method used to an-
BIOIMAGING 2025 - 12th International Conference on Bioimaging
364

alyze DTI data, focusing on the white matter tracts
in the brain. The TBSS-ENIGMA function specifi-
cally involves registering and skeletonizing fractional
anisotropy images to a common DTI atlas, allowing
for consistent and comparable analysis across differ-
ent datasets.
We carefully selected the corrected fractional
anisotropy images for each participant in our study.
These images were processed using the Tract-Based
Spatial Statistics (TBSS) pipeline developed by the
ENIGMA consortium, which encompasses several
critical preprocessing steps. Specifically, the pipeline
includes procedures for eddy current correction to
minimize distortions in the diffusion-weighted im-
ages, masking to isolate brain structures of interest,
tensor calculation for encoding the diffusion proper-
ties of the tissue, creation of the FA images that quan-
tify the directional coherence of water diffusion, and a
series of quality control checks to ensure the integrity
of the data. As part of the standardization process, we
projected the ENIGMA template onto the selected im-
ages (Smith et al., 2006), ensuring a consistent spatial
resolution of 1x1x1 mm. This high-resolution projec-
tion is crucial for accurate and reliable extraction of
imaging features.(Smith et al., 2006)
This process allowed us to gather data from the
regions of interest in the corpus callosum and cingu-
lum. We utilized a Python script to extract fractional
anisotropy values from the masked images for each
voxel, filtering out zero values. As a result, we ob-
tained 1,118 FA values for the cingulum and 7,318
for the corpus callosum.
We subsequently gathered the latest Mini-Mental
State Examination scores for each individual associ-
ated with the corresponding images. In our analy-
sis, we found that the average MMSE score for the
cognitively normal group was 28.78, showing a rel-
atively small variation with a standard deviation of
1.50. In contrast, the average MMSE score for the
Alzheimer’s disease group was significantly lower, at
22.4, accompanied by a standard deviation of 3.9, in-
dicating greater variability among the scores in this
group. To facilitate further analysis, we carefully
compiled all relevant data - voxelwise measurements,
MMSE scores, and demographic information such as
age and sex, along with unique subject identifiers -
into two distinct .csv files. These files were specifi-
cally organized to correspond to the corpus callosum
and cingulum regions of the brain, ensuring clear and
structured data for subsequent evaluation.
2.2 Clustering Data with
funWeightClust
We utilize funWeightClust (Anton and Smith, 2024b),
an advanced model-based clustering approach that
leverages a mixture of functional linear regression
models. This innovative method is particularly ben-
eficial as it accommodates functional multivariate re-
sponses and predictors, allowing for a more nuanced
analysis of complex data structures. By incorporat-
ing functional data analysis techniques, funWeight-
Clust enables us to capture the inherent variability and
relationships within multivariate datasets more effec-
tively. This capability is crucial for accurately identi-
fying clusters in situations where traditional methods
may struggle to account for the multidimensional na-
ture of the data.
The voxelwise data and MMSE scores are rep-
resented as functions, making model-based cluster-
ing methods challenging to apply; this is because
the notion of a probability density function gener-
ally does not exist for functional data (Delaigle and
Hall, 2010). FunWeightClust employs a two-step ap-
proach: first, the functional data is decomposed into
a basis of functions, and then a probabilistic model
is constructed for the coefficients of these basis func-
tions. B-spline functions are mainly used for smooth
curves, but Fourier bases are preferred for data that
exhibit a repetitive pattern. (Schmutz et al., 2020).
We aim to cluster the n observed response and pre-
dictor curves {(y
1
, x
1
), . . . , (y
n
, x
n
)} into K homoge-
neous groups. In this context, the response curves
y
i
represent the MMSE scores, while the predictor
curves x
i
are derived from the voxelwise data. For
each pair of curves (Y
i
, X
i
), we assume that these
curves belong to a finite-dimensional space, and we
have:
Y
i
(t) =
R
Y
∑
r=1
c
Y,ir
ξ
Y,r
(t), X
i
(t) =
R
X
∑
r=1
c
X,ir
ξ
X,r
(t). (1)
Here {ξ
Y,r
}
1≤r≤R
Y
and {ξ
X,r
}
1≤r≤R
X
are the
bases, R
Y
and R
X
are the number of basis func-
tions, and c
Y,ir
, c
X,ir
are the coefficients for the curves
{Y
1
, . . . , Y
n
} and the curves {X
1
, . . . , X
n
}, respec-
tively.
We assume that for each cluster k ∈ {1, . . . , K},
the observations come from the following functional
regression model:
Y
i
(t) = β
k
0
(t)+
Z
T
X
β
k
(t, s)X
i
(s)ds +E
k
(t), t ∈ T
Y
,
(2)
where i = 1, . . . , n, β
k
0
(t), β
k
(t, s) are the regression
coefficients, and E
k
(t) is the random error process
Preliminary Results on Using Clustering of Functional Data to Identify Patients with Alzheimer’s Disease by Analyzing Brain MRI Scans
365

which is uncorrelated with X
i
(s) for any (s, t) ∈ T
X
×
T
Y
.
The method funWeightClust builds upon the ap-
proach used in funHDDC (Schmutz et al., 2020). It is
based on multivariate functional principal component
analysis (MFPCA) (Jacques and Preda, 2014), assum-
ing that the scores follow multivariate normal dis-
tributions. To address regression relationships, fun-
WeightClust also incorporates extensions of cluster-
weighted models used for multivariate data (Dang
et al., 2017). Additionally, funWeightClust offers sev-
eral parsimonious models.
The Expectation-Maximization (EM) algorithm
estimates the model’s parameters. The number of
clusters is denoted as K, and the parsimonious model
is selected by maximizing the Bayesian Information
Criterion (BIC) (Schwarz, 1978). As in the case of
funHDDC, the group-specific dimension d
k
is deter-
mined using the Cattell scree test, which compares
the differences between eigenvalues against a speci-
fied threshold ε (Bouveyron and Jacques, 2011).
When the true classifications are known, the Cor-
rect Classification Rate (CCR) and the Adjusted Rand
Index (ARI) are used to measure the accuracy of the
classification. The CCR represents the ratio of cor-
rectly classified observations to the total number of
observations. The ARI adjusts for variations in clus-
ter sizes, resulting in an expected value of 0 and a
perfect classification value of 1.
3 PRELIMINARY RESULTS
We conducted a series of experiments using the R
programming language implementation of the fun-
WeightClust method. In our experiments, we applied
the functional clustering method with the number of
cluster parameters set to two. This configuration al-
lowed us to partition the data effectively into two dis-
tinct groups: corresponding to the AD and CN sub-
jects. We focused on exploring Fourier bases of vary-
ing sizes - specifically, 20, 30, and 50 - due to the
repetitive patterns observed in our data. We selected
a Fourier basis size of 50 because it provided the best
approximation of all initial curves. For the parameter
ε, we experimented with values of 0.4, 0.2, 0.1, 0.05,
0.01, 0.005, and 0.001. Using the Bayesian Informa-
tion Criterion, which is a criterion for model selec-
tion among a finite set of models, we determined that
ε = 0.001 was the most appropriate value. The BIC
helps in selecting the model that best balances com-
plexity and goodness of fit.
Our analysis included several datasets, specifi-
cally targeting the corpus callosum data, the cingulum
data, and the results from a combined dataset com-
prised of both ROI’s data sets. Through this approach,
we aimed to uncover meaningful insights from the un-
derlying structures in the data.
We conducted two sets of experiments: Experi-
ment A, which included only the voxelwise data, and
Experiment B, which incorporated the voxelwise data
along with age as input variables. In Experiment A,
the results of funWeightClust for the cingulum and
corpus callosum data varied based on the values of ε.
The most favorable outcomes were achieved using a
Fourier base of size 50 with ε = 0.001, as shown in ta-
bles 1, 2 and 3. For the combined data in Experiment
A, the results remained consistent across all ε values.
Experiment B yielded similar results. Including
age data resulted in a slight improvement in the ad-
justed Rand index while causing a marginal decrease
in the correct classification rate. For instance, using
a Fourier base of size 50 with ε = 0.001 for the com-
bined cingulum and corpus callosum datasets, Exper-
iment B produced an ARI of 0.6132 and a CCR of
0.7954. In this preliminary investigation, we priori-
tized Experiment A because the improvements in Ex-
periment B were too small.
For comparison purposes, we also performed sim-
ilar experiments using funHDDC (Schmutz et al.,
2020) and tFunHDDC (Anton and Smith, 2024a).
funHDDC (Functional High-Dimensional Data Clus-
tering) is a method designed to cluster high-
dimensional functional data, while tFunHDDC is an
extension that uses t-distribution instead of a normal
distribution. Both funHDDC and tfunHDDC do not
include a linear regression relationship, so we ran
these methods only on the predictor curves derived
from the voxelwise data. We used the same setup for
the dataset and parameters, specifically the size of the
Fourier basis and the value of ε.
The results of Experiment A, funHDDC and tfun-
HDDC for the cingulum, the corpus callosum and the
combination of the two ROIs are presented in tables
1, 2 and 3, respectively.
Taking into account the conclusions of (Ma et al.,
2019), we performed a variation of Experiment A,
where we filtered the cingulum data to contain only
the Left Hippocampus and the corpus callosum data
to contain only the Splenium. The best results were
obtained with a Fourier base of size 30 and are pre-
sented in Table 4. The ε values did not influence the
results.
Our proposed approach successfully identifies pa-
tients with Alzheimer’s disease. We believe that in-
corporating additional biomarkers could further en-
hance these results.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
366
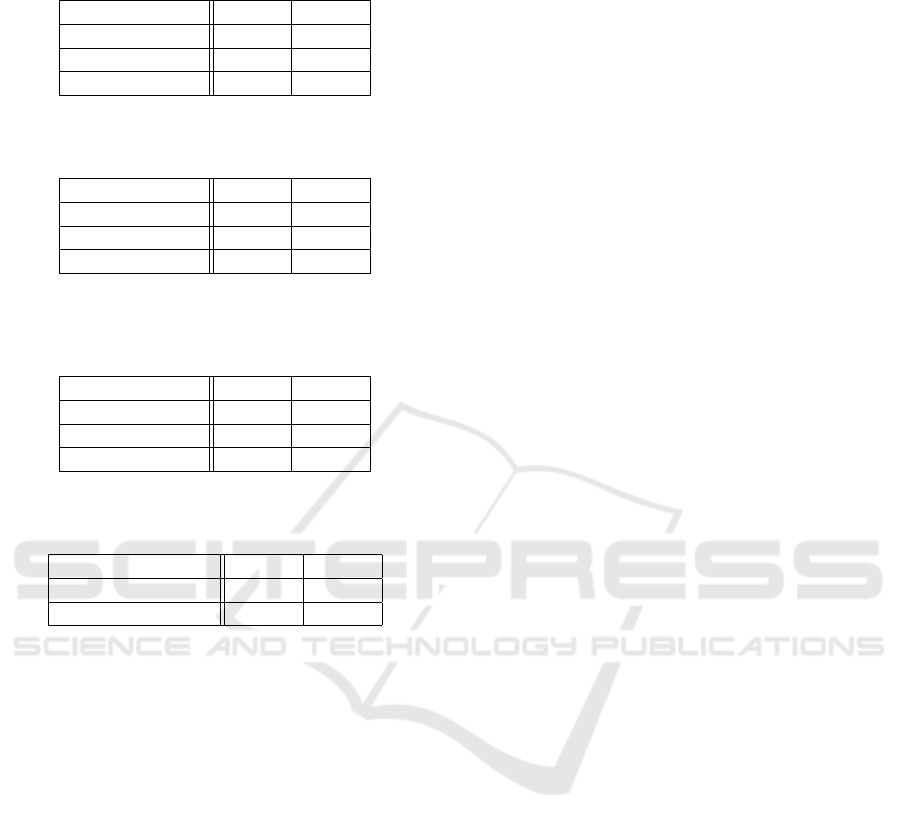
Table 1: Results for the Cingulum ROI dataset for Fourier
base of size 50 and ε = 0.001.
Method ARI CCR
funWeightClust 0.5737 0.8820
funHDDC 0.1299 0.6839
tfunHDDC 0.1163 0.6745
Table 2: Results for the Corpus Callosum ROI dataset for
Fourier base of size 50 and ε = 0.001.
Method ARI CCR
funWeightClust 0.5737 0.8820
funHDDC 0.0978 0.6603
tfunHDDC 0.1163 0.6745
Table 3: Results for the combination of the Cingulum and
the Corpus Callosum ROI datasets for Fourier base of size
50 and ε = 0.001.
Method ARI CCR
funWeightClust 0.5452 0.8726
funHDDC 0.1159 0.6698
tfunHDDC 0.1163 0.6745
Table 4: Experiment A results for selected regions. Fourier
base of size 30 and ε = 0.001.
Region ARI CCR
Left Hippocampus 0.7014 0.9198
Splenium 0.5606 0.8773
4 CONCLUSIONS
This initial study highlights the potential of fun-
WeightClust, a model-based clustering method that
employs functional linear regression models for iden-
tifying patients with Alzheimer’s disease. By an-
alyzing voxelwise fractional anisotropy data from
the ADNI dataset, we successfully distinguished be-
tween the Alzheimer’s disease and cognitively normal
groups.
The inclusion of age data slightly improved the
adjusted Rand index while causing a marginal de-
crease in the correct classification rate. This sug-
gests that demographic factors can enhance cluster-
ing accuracy. Further analysis of specific brain re-
gions, such as the left hippocampus and the splenium,
yielded better results. Our preliminary findings indi-
cate that funWeightClust is a promising tool for the
early detection of Alzheimer’s disease. The proposed
new functional clustering method demonstrates better
clustering results than other existing similar methods.
A key strength of funWeightClust lies in its flexibil-
ity with response variables. Unlike our preliminary
study, where we used only MMSE as the response
variable, this new approach allows for the extension
to functional values. This means that the response can
be a vector of multiple scores, such as a combination
of MMSE, Montreal Cognitive Assessment (MoCA),
and auditory verbal learning test (AVLT), thus provid-
ing a more comprehensive and nuanced analysis. This
capability enhances the method’s applicability and ef-
fectiveness, making it a significant advancement. Fu-
ture research should focus on expanding the dataset
and incorporating additional biomarkers to further en-
hance the method’s effectiveness, thereby improving
early detection techniques for AD.
ACKNOWLEDGMENTS
Data collection and sharing for this project was
funded by the Alzheimer’s Disease Neuroimaging
Initiative (ADNI) (National Institutes of Health Grant
U01 AG024904) and DOD ADNI (Department of De-
fense award number W81XWH-12-2-0012). ADNI
is funded by the National Institute on Aging, the
National Institute of Biomedical Imaging and Bio-
engineering, and through generous contributions from
the following: AbbVie, Alzheimer’s Association;
Alzheimer’s Drug Discovery Foundation; Araclon
Biotech; BioClinica, Inc.; Biogen; Bristol-Myers
Squibb Company; CereSpir, Inc.; Cogstate; Eisai
Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Com-
pany; EuroImmun; F. Hoffmann-La Roche Ltd and
its affiliated company Genentech, Inc.; Fujirebio;
GE Healthcare; IXICO Ltd.; Janssen Alzheimer Im-
munotherapy Research & Development, LLC.; John-
son & Johnson Pharmaceutical Research & Devel-
opment LLC.; Lumosity; Lundbeck; Merck & Co.,
Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Re-
search; Neurotrack Technologies; Novartis Pharma-
ceuticals Corporation; Pfizer Inc.; Piramal Imag-
ing; Servier; Takeda Pharmaceutical Company; and
Transition Therapeutics. The Canadian Institutes of
Health Research is providing funds to support ADNI
clinical sites in Canada. Private sector contributions
are facilitated by the Foundation for the National In-
stitutes of Health (http://www.fnih.org). The grantee
organization is the Northern California Institute for
Research and Education, and the study is coordinated
by the Alzheimer’s Therapeutic Research Institute at
the University of Southern California. ADNI data are
disseminated by the Laboratory for Neuro Imaging at
the University of Southern California.
The second author was supported by the Natu-
ral Sciences and Engineering Research Council of
Preliminary Results on Using Clustering of Functional Data to Identify Patients with Alzheimer’s Disease by Analyzing Brain MRI Scans
367

Canada (NSERC) through the grant DG-2018-04449.
The work of the fourth author was supported by
an Alberta Innovates Summer Research Studentship.
The work of the fifth author was supported by an
NSERC USRA grant.
REFERENCES
Anton, C. and Smith, I. (2024a). Model-based clus-
tering of functional data via mixtures of t distribu-
tions. Advances in Data Analysis and Classification,
18(3):563–595.
Anton, C. and Smith, I. (2024b). A multivariate functional
data clustering method using parsimonious cluster
weighted models. In Theodore Chadjipadelis, Au-
rea Gran
´
e, J. T. and Villalobos, M., editors, Data
Science, Classification and Artificial Intelligence for
Modeling Decision Making, Studies in Classifica-
tion, Data Analysis, and Knowledge Organization.
Springer International Publishing. to appear.
Armstrong, J. J., Guimond, J., Sandals, L., Neufeld, B.,
Christie, N., Perry, S., John, J., Akintade, T., and
Bayne, S. (2022). Navigating the path forward for de-
mentia in Canada.
Bouveyron, C. and Jacques, J. (2011). Model-based clus-
tering of time series in group-specific functional sub-
spaces. Adv Data Anal Classif., 5(4):281–300.
Dang, U. J., Punzo, A., McNicholas, P. D., Ingrassia, S.,
and Browne, R. P. (2017). Multivariate response and
parsimony for Gaussian cluster-weighted models. J.
Classif., 34(1):4–34.
Dara, O. A., Lopez-Guede, J. M., Raheem, H. I., Ra-
hebi, J., Zulueta, E., and Fernandez-Gamiz, U.
(2023). Alzheimer’s Disease Diagnosis Using Ma-
chine Learning: A Survey. Applied Sciences, 13(14).
Delaigle, A. and Hall, P. (2010). Defining probability den-
sity for a distribution of random functions. Ann. Stat.,
38(2):1171–1193.
Jacques, J. and Preda, C. (2014). Model-based clustering
for multivariate functional data. Computational Statis-
tics & Data Analysis, 71:92–106.
Jahanshad, N., Kochunov, P., Sprooten, E., Mandl, R.,
Nichols, T., Almassy, L., Blangero, J., Brouwer, R.,
Curran, J., de Zubicaray, G., Duggirala, R., Fox, P.,
Hong, L., Landman, B., Martin, N., McMahon, K.,
Medland, S., Mitchell, B., Olvera, R., and Glahn, D.
(2013). Multi-site genetic analysis of diffusion images
and voxelwise heritability analysis: A pilot project of
the enigma-dti working group. NeuroImage, 81.
Kaur, A., Mittal, M., Bhatti, J. S., Thareja, S., and Singh, S.
(2024). A systematic literature review on the signif-
icance of deep learning and machine learning in pre-
dicting alzheimer’s disease. Artificial Intelligence in
Medicine, 154:102928.
Li, T., Song, X., Zhang, Y., Zhu, H., and Zhu, Z.
(2021). Clusterwise functional linear regression
models. Computational Statistics & Data Analysis,
158:107192.
Ma, H., Li, T., Zhu, H., and Zhu, Z. (2019). Quantile regres-
sion for functional partially linear model in ultra-high
dimensions. Computational Statistics & Data Analy-
sis, 129:135–147.
Nir, T. M., Jahanshad, N., Villalon-Reina, J. E., Toga,
A. W., Jack, C. R., Weiner, M. W., and Thompson,
P. M. (2013). Effectiveness of regional DTI measures
in distinguishing Alzheimer’s disease, MCI, and nor-
mal aging. NeuroImage: Clinical, 3:180–195.
Oishi, K., Mielke, M. M., Albert, M., Lyketsos, C. G., and
Mori, S. (2011). DTI analyses and clinical applica-
tions in Alzheimer’s disease. Journal of Alzheimer’s
Disease, 26(s3):287–296.
Schmutz, A., Jacques, J., Bouveyron, C., Cheze, L., and
Martin, P. (2020). Clustering multivariate functional
data in group-specific functional subspaces. Comput.
Stat., 35:1101–1131.
Schouten, T., Koini, M., de Vos, F., Seiler, S., Rooij, M.,
Lechner, A., Schmidt, R., Heuvel, M., van der Grond,
J., and Rombouts, S. (2017). Individual Classification
of Alzheimer’s Disease with Diffusion Magnetic Res-
onance Imaging. NeuroImage, 152.
Schwarz, G. (1978). Estimating the dimension of a model.
Ann. Stat., pages 461–464.
Smith, S. M., Jenkinson, M., Johansen-Berg, H., Rueck-
ert, D., Nichols, T. E., Mackay, C. E., Watkins, K. E.,
Ciccarelli, O., Cader, M. Z., Matthews, P. M., and
Behrens, T. E. (2006). Tract-based spatial statistics:
Voxelwise analysis of multi-subject diffusion data.
NeuroImage, 31(4):1487–1505.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
368
