
Leveraging Large Language Models and RNNs for Accurate
Ontology-Based Text Annotation
Pratik Devkota
1 a
, Somya D. Mohanty
2 b
and Prashanti Manda
3 c
1
Informatics and Analytics, University of North Carolina, Greensboro, NC, U.S.A.
2
United Health Group, U.S.A.
3
Department of Computer Science, University of Nebraska Omaha, NE, U.S.A.
Keywords:
Ontology Annotation, Large Language Models, Gene Ontology, Biomedical NLP, Model Fine-Tuning.
Abstract:
This study investigates the performance of large language models (LLMs) and RNN-based architectures for
automated ontology annotation, focusing on Gene Ontology (GO) concepts. Using the Colorado Richly Anno-
tated Full-Text (CRAFT) dataset, we evaluated models across metrics such as F1 score and semantic similarity
to measure their precision and understanding of ontological relationships. The Boosted Bi-GRU, a lightweight
model with only 38M parameters, achieved the highest performance, with an F1 score of 0.850 and semantic
similarity of 0.900, demonstrating exceptional accuracy and computational efficiency. In comparison, LLMs
like Phi (1.5B) performed competitively, balancing moderate GPU usage with strong annotation accuracy.
Larger models, including Mistral, Meditron, and Llama 2 (7B), delivered comparable results but required sig-
nificantly higher computational resources for fine-tuning and inference, with GPU usage exceeding 125 GB
during fine-tuning. Fine-tuned ChatGPT 3.5 Turbo underperformed relative to other models, while ChatGPT
4 showed limited applicability for this domain-specific task. To enhance model performance, techniques such
as prompt tuning and full fine-tuning were employed, incorporating hierarchical ontology information and
domain-specific prompts. These findings highlight the trade-offs between model size, resource efficiency, and
accuracy in specialized tasks. This work provides insights into optimizing ontology annotation workflows and
advancing domain-specific natural language processing in biomedical research.
1 INTRODUCTION
Automatically annotating scientific literature with do-
main ontology concepts is crucial in fields like biol-
ogy and biomedical sciences (Dahdul et al., 2015).
This process involves tagging and linking text to
predefined ontologies using NLP techniques (Manda
et al., 2020), enabling structured knowledge extrac-
tion from unstructured text.
Ontology annotation aids in knowledge manage-
ment, literature review, data integration, and appli-
cations like information retrieval, knowledge graphs,
and semantic search. The growth of biological on-
tologies has driven research into NLP methods for
automating this task (Devkota et al., 2022b; Devkota
et al., 2022a). This enhances information organiza-
tion and connects related research effectively.
a
https://orcid.org/0000-0001-5161-0798
b
https://orcid.org/0000-0002-4253-5201
c
https://orcid.org/0000-0002-7162-7770
Automated ontology annotation involves several
key steps. It begins with text processing, where the
literature is preprocessed to clean and prepare the text
for analysis. This is followed by entity recognition,
which identifies significant entities or terms within the
text. These recognized entities are then mapped to
corresponding concepts in the ontology through on-
tology mapping. Once the mapping is established, an-
notations are added to the text in the form of metadata
or tags. Finally, the process concludes with validation
to ensure the annotations are accurate and relevant.
Traditional machine learning methods like RNNs
and CNNs have been widely used for automated
ontology annotation of scientific literature (Lample
et al., 2016; Boguslav et al., 2021; Casteleiro et al.,
2018; Manda et al., 2020; Devkota et al., 2023; De-
vkota et al., 2022a). Our team has employed Bi-
GRUs, leveraging their sequential processing capa-
bilities to enhance performance on ontology annota-
tion tasks (Manda et al., 2018; Manda et al., 2020;
Devkota et al., 2022b; Devkota et al., 2022a; De-
Devkota, P., Mohanty, S. D. and Manda, P.
Leveraging Large Language Models and RNNs for Accurate Ontology-Based Text Annotation.
DOI: 10.5220/0013267100003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 489-494
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
489

vkota et al., 2023; Pratik et al., 2023). A GRU-based
model focusing on extracting Gene Ontology (GO)
terms demonstrated strong results using ELMo em-
beddings for better contextual understanding, achiev-
ing high F1 scores and Jaccard similarity (Devkota
et al., 2022b). This approach addresses the complex-
ity of biomedical texts, where concepts are often in-
directly implied.
We introduced an ontology-aware annotation ap-
proach for biological literature (Devkota et al., 2022a)
that leverages hierarchical and semantic relation-
ships in structured ontologies like Gene Ontology
(GO). By integrating these relationships into training,
the model distinguishes related terms and captures
context-specific meanings more effectively. Using
embeddings like CRAFT, GloVe, and ELMo, the ap-
proach improved performance by up to 10%, achiev-
ing higher F1 scores and Jaccard similarity through
enhanced semantic accuracy.
Enhancing GRU-based architectures with a post-
processing technique that leveraged structured on-
tologies significantly improved semantic understand-
ing and annotation accuracy (Devkota et al., 2023).
By incorporating hierarchical relationships, such as
those in the Gene Ontology, the model captured nu-
anced term connections and used semantic similar-
ity metrics to address concept variability and indirect
references. This resulted in more accurate, context-
sensitive annotations, improving literature mining in
complex biomedical texts.
Previous work trained neural networks to map
words in a gold-standard corpus to ontology concepts,
achieving state-of-the-art annotation with low mem-
ory use and fast inference. With advancements in
Large Language Models (LLMs), the question arises:
can LLMs improve ontology annotation, and is their
higher computational cost justified?
Large language models (LLMs), like OpenAI’s
GPT series and Google’s BERT, use transformer ar-
chitectures to process and generate human language
based on vast text datasets (Vaswani, 2017). These
models have been widely adopted for tasks like con-
tent creation and customer service due to their abil-
ity to perform various language tasks with minimal
fine-tuning (Brown, 2020). However, they have limi-
tations, such as producing inaccurate “hallucinations”
and relying heavily on the quality of their training
data (Bender et al., 2021). Research is focused on im-
proving their factual accuracy and energy efficiency,
given their high computational demands (Strubell
et al., 2020).
This study explores the use of LLMs for au-
tomated ontology annotation of scientific literature,
with a focus on Gene Ontology (GO) annotations.
Model performance was assessed using metrics like
F1 score and semantic similarity, evaluating their se-
mantic accuracy in annotations. This work aims to
improve the performance of large language models
(LLMs) for ontology annotation in scientific litera-
ture, focusing on the Gene Ontology (GO). It in-
volves experimenting with models like MPT-7B, Phi,
BiomedLM, and Meditron to determine which best
capture complex semantic relationships in ontology-
based text.
2 METHODS
2.1 Dataset
The Colorado Richly Annotated Full-Text (CRAFT)
dataset, an annotated corpus of 97 full-text biomedi-
cal articles, was used to train the models. Covering
domains like Gene Ontology (GO), ChEBI, and Se-
quence Ontology (SO), it provides detailed annota-
tions for tasks such as named entity recognition, on-
tology mapping, and semantic analysis, making it es-
sential for biomedical text analysis.
The CRAFT corpus was segmented into 27,946
sentences, each containing zero or more words or
phrases annotated with unique Gene Ontology (GO)
IDs. The dataset was divided into 22,364 training sen-
tences and 5,582 evaluation sentences. This frame-
work was used to evaluate and optimize large lan-
guage models (LLMs) for accurately predicting GO
concepts linked to words or phrases in input sen-
tences.
2.2 Baseline Model Selection and
Comparison Framework
In prior work, we trained Bi-GRU models on the
CRAFT dataset, enhanced with parts-of-speech tags
and data from NCBI’s BioThesaurus and UMLS. The
best model achieved an F1 score and semantic sim-
ilarity of 0.84, serving as a baseline for comparing
fine-tuned large language models (LLMs) on the same
dataset.
We developed a post-processing technique called
“Ontology Boosting” (Devkota et al., 2023) to en-
hance the confidence of predictions from Bi-GRU
models, achieving an F1 score of 0.85 and a semantic
similarity of 0.90. During LLM fine-tuning experi-
ments, we will compare their performance and mem-
ory efficiency against our Bi-GRU baseline.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
490

2.3 Large Language Models
For our experiments, we selected MPT-7B, a
seven-billion-parameter decoder-style transformer
pretrained on one trillion English text and code to-
kens, as the foundational LLM. Its manageable size
and efficient training and inference throughput made
it ideal for balancing performance with computational
efficiency. This choice ensured a fair comparison of
RNNs and LLMs in terms of both performance and
resource usage.
We also selected the following models for com-
parison with our baseline model:
2.3.1 Phi
Phi, developed by Google DeepMind, enhances trans-
former models for complex reasoning, particularly in
multi-step tasks. It delivers high-quality responses
across topics like scientific research and general
knowledge, leveraging advanced techniques to learn
from both structured and unstructured data for deeper
understanding and contextual sensitivity.
2.3.2 BiomedLM
BiomedLM is a domain-specific LLM fine-tuned on
biomedical literature, clinical reports, and medical
datasets, delivering accurate outputs in medical con-
texts. It excels in tasks like drug discovery, bioin-
formatics, medical research, and clinical decision
support, thanks to its deep understanding of com-
plex biomedical terminologies and relationships. This
makes it a valuable tool for healthcare profession-
als navigating medical knowledge and generating in-
sights for new discoveries.
2.3.3 Falcon
Falcon, developed by the Technology Innovation In-
stitute, is an efficient large language model optimized
for generative and analytical tasks. It delivers coher-
ent, contextually accurate responses with low compu-
tational demands, making it ideal for real-world appli-
cations in resource-constrained settings. Excelling in
text summarization, question-answering, and natural
language generation, Falcon balances speed and accu-
racy, enabling its use across industries like healthcare
and e-commerce.
2.3.4 Meditron
Meditron is a healthcare-focused LLM fine-tuned for
processing medical texts and clinical data. Opti-
mized for understanding complex medical terminol-
ogy, it supports tasks like diagnosis assistance, clin-
ical decision-making, and patient care recommenda-
tions, ensuring high accuracy in critical medical con-
texts.
2.3.5 Llama 2
Llama 2, developed by Meta, is a versatile LLM op-
timized for general NLP tasks like text generation,
translation, summarization, and question-answering.
Its scalable design ensures high performance and
adaptability for both research and commercial use.
2.3.6 Mistral
Mistral is an open-weight, high-performance LLM
designed for multitask learning and fine-tuning in do-
mains like programming, healthcare, and customer
service. It efficiently adapts to diverse tasks and
datasets without extensive retraining.
2.3.7 MPT
MPT (Mosaic Pretrained Transformer) by MosaicML
is an open-source, efficient LLM optimized for tasks
like text generation, summarization, and question-
answering. Its scalability and adaptability make it
ideal for industries like finance, healthcare, and ed-
ucation, offering cost-effective fine-tuning on smaller
datasets.
2.3.8 Finetuned ChatGPT
Finetuned ChatGPT refers to customized versions of
OpenAI’s GPT models, optimized for specific tasks
or datasets. While the base model excels in general-
purpose applications, fine-tuning enhances its accu-
racy and relevance in specialized domains, improving
performance in targeted conversational AI tasks.
2.4 Fine-Tuning for the Initial Model
We carried out a comprehensive fine-tuning process
for the initial model, divided into four distinct stages:
2.4.1 Prompt Tuning
We initiated the prompt-tuning stage to improve gen-
erative performance and minimize hallucinations in
the fine-tuned model. This began with a single task,
instructing the model to extract terms linked to GO
concepts from input sentences. The prompt required
the model to identify and extract words or phrases re-
lated to the GO hierarchy or indicate if no associa-
tions were found. An example of the initial prompt-
response data is shown below:
Prompt:
Leveraging Large Language Models and RNNs for Accurate Ontology-Based Text Annotation
491

Instruction: Use the input sentence below to ex-
tract terms that are associated with some concept in
Gene Ontology hierarchy.
Input: Interactions of CSS for arterial thrombus
formation
Response: Terms: thrombus formation
This prompt-response format served as the foun-
dation for fine-tuning, creating a dataset applied to
both training and evaluation. The fine-tuned model
used prompts to generate responses based on learned
GO associations, which were compared to ground
truth annotations for performance assessment.
We refined the prompts iteratively, adjusting lan-
guage and specificity for greater accuracy. Initially,
prompts included GO IDs, but this caused halluci-
nations with invalid IDs. Removing IDs and instead
instructing the model to include parent concepts im-
proved its understanding of the ontology hierarchy.
Contextualizing prompts as if from a gene ontol-
ogy expert further enhanced relevance and coherence,
guiding the model to focus on domain-specific terms.
Formatting adjustments, such as JSON outputs
and uppercase keywords, improved clarity and post-
processing, enhancing the structure and usability of
generated responses. These iterative changes culmi-
nated in a final prompt design instructing the model
to associate concepts, include parent terms, and adopt
the persona of a gene ontology expert, optimizing its
performance in generating accurate ontology annota-
tions.
2.4.2 Architecture Tuning
After finalizing the optimal prompt, we proceeded to
the next phase, exploring supervised fine-tuning tech-
niques to further train the pretrained large language
model on our smaller dataset. This aimed to enhance
the model’s performance in ontology annotation. We
focused on full fine-tuning in this study. Full fine-
tuning involved training the entire model, including
all layers and parameters, for the ontology annotation
task. Using the final prompt template, we determined
a maximum sequence length of 1024 tokens to bal-
ance dataset coverage and memory efficiency during
training and inference.
Extensive experimentation optimized fine-tuning
parameters, yielding the best results with a batch size
of 8, 3 training epochs, a learning rate of 5.0e-06,
and the decoupled AdamW optimizer with linear de-
cay and 50 warm-up batches. To improve compu-
tational efficiency, we leveraged flash attention for
faster and memory-efficient operations and employed
Full Sharded Data Parallel (FSDP) to shard opti-
mizer states, gradients, and parameters across work-
ers. These techniques enabled training of the 7-
billion-parameter MPT model with a global batch size
of 24 on 3 NVIDIA A6000 GPUs (48GB each).
2.5 Performance Evaluation Metrics
The performance of the baseline and boosted Bi-GRU
models was evaluated using a modified F1 score and
Jaccard semantic similarity. The modified F1 ex-
cluded accurately predicted out-of-concept tokens to
minimize bias, as these tokens, unrelated to specific
concepts, were abundant in the dataset. In contrast,
LLMs, which generate text rather than predict indi-
vidual tokens, were evaluated using the unmodified
F1 score and Jaccard semantic similarity (Pesquita
et al., 2009).
The F1 score measured precise concept annota-
tion, while the Jaccard semantic similarity assessed
the ontological distance between annotated concepts,
evaluating the model’s ability to provide semantically
similar alternatives when exact matches were miss-
ing. This offered insights into the model’s semantic
understanding of the ontology.
3 RESULTS
We compared the performance of our Bi-GRU base-
line model with various LLMs using F1 Score and
Semantic Similarity Score (Figure 1). Model sizes
ranged from 38 million to several billion parame-
ters. Boosted Bi-GRU (38M) and Phi (1.5B) achieved
the highest performance, with Boosted Bi-GRU scor-
ing 0.850 in F1 and 0.900 in semantic similarity, ex-
celling in semantic understanding despite its small
size. Larger models, including Mistral, Meditron, and
Llama 2 (all 7B), showed similar performance, with
F1 scores between 0.839 and 0.878 and semantic sim-
ilarity scores from 0.840 to 0.876. Fine-tuned Chat-
GPT 3.5 Turbo (3.5B) scored lower, with an F1 of
0.685 and semantic similarity of 0.699. ChatGPT 4
performed the worst, with an F1 of 0.048 and seman-
tic similarity of 0.061, indicating significant under-
performance in this context.
We compared GPU usage across models during
finetuning and inference (Figure 2), measured in giga-
bytes (GB). Light green bars represent finetuning us-
age, while dark green bars show inference usage. The
7B models—Falcon, Meditron, Llama 2, Mistral, and
MPT—had the highest GPU usage during finetuning,
ranging from 125.3 GB (Llama 2) to 138.9 GB (Mis-
tral), and maintained high inference usage around 15-
16 GB.
Boosted Bi-GRU, the smallest model with 38M
parameters, was the most resource-efficient, using
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
492
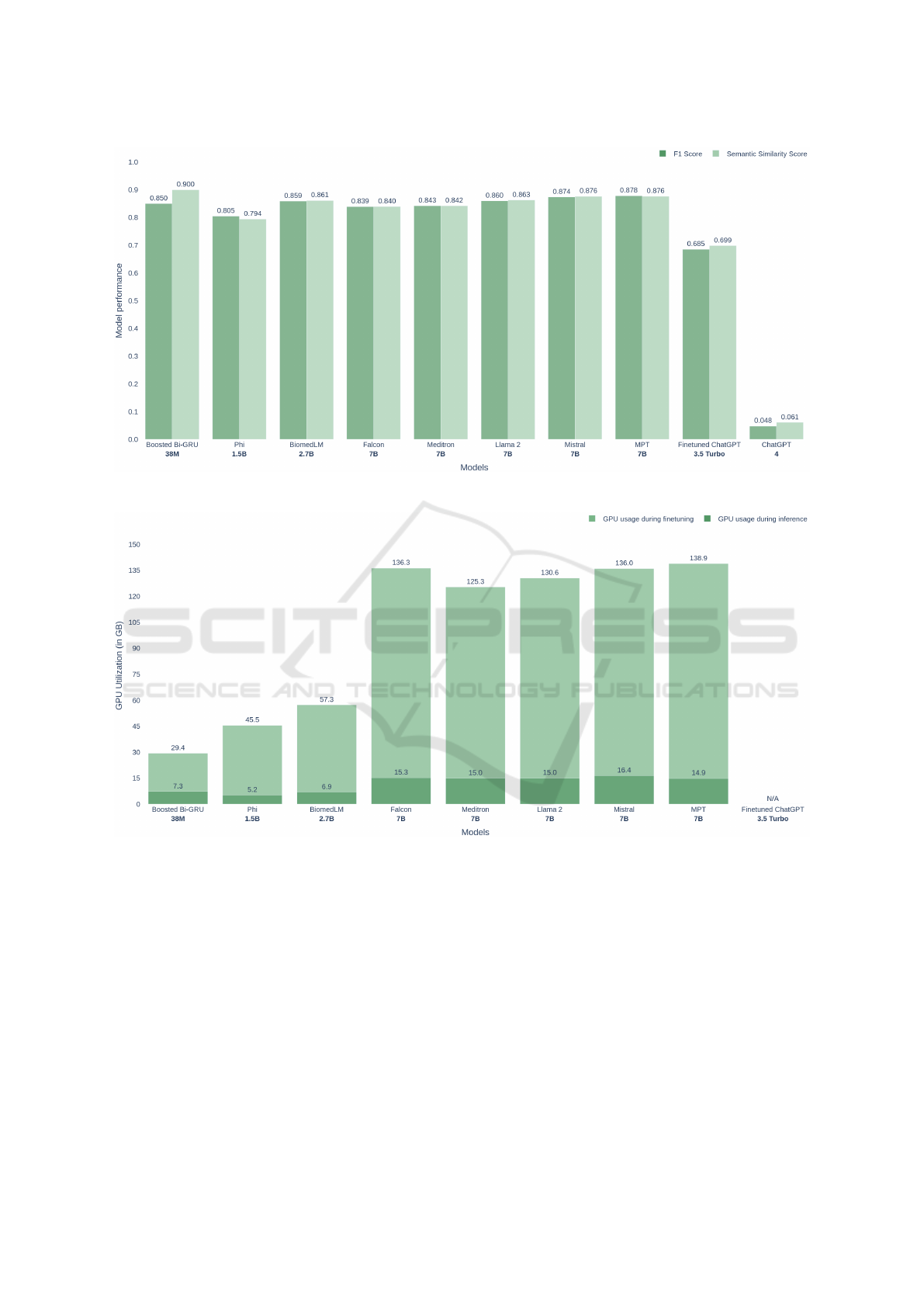
Figure 1: Performance comparison between RNN based model and different LLMs.
Figure 2: GPU utilization during finetuning and inferencing by different models.
only 29.4 GB during finetuning and 7.3 GB during in-
ference. Phi (1.5B) and BiomedLM (2.7B) had mod-
erate GPU utilization during finetuning (45.5 GB and
57.3 GB, respectively) and low inference usage (5.2
GB and 6.9 GB). Finetuned ChatGPT 3.5 Turbo’s
GPU usage data was unavailable or not applicable for
this comparison.
4 CONCLUSIONS
This study evaluated the performance and resource ef-
ficiency of various large language models (LLMs) and
RNN-based models for automated ontology annota-
tion, focusing on Gene Ontology (GO) concepts. Our
findings demonstrated that smaller models like the
Boosted Bi-GRU, despite its modest 38M parameters,
achieved remarkable semantic understanding with an
F1 score of 0.850 and a semantic similarity score of
0.900, outperforming or matching larger LLMs in ac-
curacy while being highly resource-efficient.
Among the LLMs, Phi (1.5B) exhibited competi-
tive performance, combining strong semantic under-
standing with moderate resource usage. Larger mod-
els like Mistral, Meditron, and Llama 2 (7B) showed
comparable annotation quality but required signifi-
Leveraging Large Language Models and RNNs for Accurate Ontology-Based Text Annotation
493

cantly higher GPU resources for fine-tuning and in-
ference. Notably, ChatGPT 4 underperformed in this
task, highlighting the limitations of general-purpose
LLMs without domain-specific fine-tuning.
In terms of computational efficiency, the Boosted
Bi-GRU model demonstrated the best trade-off be-
tween accuracy and resource usage, while models like
Phi and BiomedLM provided a balance of scalabil-
ity and performance in biomedical contexts. These
findings underscore the importance of aligning model
selection and fine-tuning strategies with task-specific
requirements and resource constraints.
Future work will explore advanced parameter-
efficient fine-tuning techniques, such as adapters or
LoRA, to further enhance the capabilities of large
models while minimizing computational costs. Addi-
tionally, integrating more sophisticated semantic sim-
ilarity metrics and hierarchical context into evaluation
frameworks may yield deeper insights into model per-
formance in ontology-driven tasks. This work pro-
vides a foundation for developing scalable and accu-
rate models for ontology annotation in specialized do-
mains like biomedical sciences.
ACKNOWLEDGEMENTS
This work is funded by a CAREER award (#1942727)
from the Division of Biological Infrastructure at the
National Science Foundation, USA.
REFERENCES
Bender, E. M., Gebru, T., McMillan-Major, A., and
Shmitchell, S. (2021). On the dangers of stochastic
parrots: Can language models be too big? In Pro-
ceedings of the 2021 ACM conference on fairness, ac-
countability, and transparency, pages 610–623.
Boguslav, M. R., Hailu, N. D., Bada, M., Baumgartner,
W. A., and Hunter, L. E. (2021). Concept recognition
as a machine translation problem. BMC bioinformat-
ics, 22(1):1–39.
Brown, T. B. (2020). Language models are few-shot learn-
ers.
Casteleiro, M. A., Demetriou, G., Read, W., Prieto, M. J. F.,
Maroto, N., Fernandez, D. M., Nenadic, G., Klein,
J., Keane, J., and Stevens, R. (2018). Deep learning
meets ontologies: experiments to anchor the cardio-
vascular disease ontology in the biomedical literature.
Journal of biomedical semantics, 9(1):13.
Dahdul, W., Dececchi, T. A., Ibrahim, N., Lapp, H., and
Mabee, P. (2015). Moving the mountain: analysis of
the effort required to transform comparative anatomy
into computable anatomy. Database, 2015.
Devkota, P., Mohanty, S., and Manda, P. (2022a). Knowl-
edge of the ancestors: Intelligent ontology-aware an-
notation of biological literature using semantic simi-
larity.
Devkota, P., Mohanty, S., and Manda, P. (2023). Ontology-
powered boosting for improved recognition of on-
tology concepts from biological literature [ontology-
powered boosting for improved recognition of ontol-
ogy concepts from biological literature]. In 16th In-
ternational Joint Conference on Biomedical Engineer-
ing Systems and Technologies (BIOSTEC 2023), vol-
ume 3.
Devkota, P., Mohanty, S. D., and Manda, P. (2022b). A
gated recurrent unit based architecture for recognizing
ontology concepts from biological literature. BioData
Mining, 15(1):1–23.
Lample, G., Ballesteros, M., Subramanian, S., Kawakami,
K., and Dyer, C. (2016). Neural architectures for
named entity recognition.
Manda, P., Beasley, L., and Mohanty, S. (2018). Taking
a dive: Experiments in deep learning for automatic
ontology-based annotation of scientific literature.
Manda, P., SayedAhmed, S., and Mohanty, S. D. (2020).
Automated ontology-based annotation of scientific lit-
erature using deep learning. In Proceedings of The
International Workshop on Semantic Big Data, SBD
’20, New York, NY, USA. Association for Computing
Machinery.
Pesquita, C., Faria, D., Falcao, A. O., Lord, P., and Couto,
F. M. (2009). Semantic similarity in biomedical on-
tologies. PLoS computational biology, 5(7).
Pratik, D., Somya, D. M., and Prashanti, M. (2023). Im-
proving the evaluation of nlp approaches for scien-
tific text annotation with ontology embedding-based
semantic similarity metrics. In Proceedings of the
20th International Conference on Natural Language
Processing (ICON), pages 516–522.
Strubell, E., Ganesh, A., and McCallum, A. (2020). Energy
and policy considerations for modern deep learning
research.
Vaswani, A. (2017). Attention is all you need.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
494
