
De-Anonymization of Health Data: A Survey of Practical Attacks,
Vulnerabilities and Challenges
∗
Hamza Aguelal
a
and Paolo Palmieri
b
Department of Computing & IT, University College Cork, Cork, Ireland
Keywords:
De-Anonymization Attacks, Anonymity, Anonymization Assessment, Health Data Protection, Data Privacy.
Abstract:
Health data ranks among the most sensitive personal information disclosing serious details about individu-
als. Although anonymization is used, vulnerabilities persist, leading to de-anonymization and privacy risks
highlighted by regulations like the General Data Protection Regulation (GDPR). This survey examines de-
anonymization attacks on health datasets, focusing on methodologies employed, data targeted, and the effec-
tiveness of current anonymization practices. Unlike previous surveys that lack consensus on essential empirical
questions, we provide a comprehensive summary of practical attacks, offering a more logical perspective on
real-world risk. Our investigation systematically categorizes these practical attacks, revealing insights into suc-
cess rates, generality and reproducibility, new analytics used, and the specific vulnerabilities they exploit. The
study covers health-related datasets, including medical records, genomic data, electrocardiograms (ECGs),
and neuroimaging, highlighting the need for more robust anonymization. Significant challenges remain in
the literature despite existing reviews. We advocate for stronger data safeness by improving anonymization
methods and advancing research on de-anonymization and assessment within healthcare.
1 INTRODUCTION
Digitalization of healthcare raised challenges, threats,
and complexity in safeguarding patient privacy, espe-
cially with advancements in medical research, public
health, and personalized medicine. As highly sensi-
tive, data misuse can lead to severe privacy breaches
and risks, particularly de-anonymization. Regula-
tions like GDPR (European Union, 2016) and the
Health Insurance Portability and Accountability Act
(HIPAA) (U.S. Congress, 1996) emphasize the need
for stringent data protection in healthcare.
Despite advancements in anonymization prac-
tices, healthcare remains a prime target for breaches,
per IBM’s report (IBM Security, 2023). We highlight
a
https://orcid.org/0009-0002-9409-4206
b
https://orcid.org/0000-0002-9819-4880
∗
Funded in part by the European Union (EU),
Grant Agreement no.101095717 (SECURED) and by
Taighde
´
Eireann – Research Ireland under Grant number
18/CRT/6222. For the purpose of Open Access, the author
has applied a CC BY public copyright licence to any Author
Accepted Manuscript version arising from this submission.
Views and opinions expressed are those of the authors and
do not necessarily reflect those of the EU or the Health and
Digital Executive Agency. Neither the EU nor the granting
authority are responsible for them.
insufficiency against vulnerabilities exploited via ad-
vanced analytics and auxiliary information.
Previous studies have explored de-anonymization
techniques, but there is a noticeable gap focusing on
practical attacks in healthcare; many address theo-
retical risks in other fields (different to healthcare).
This hinders the development of effective counter-
measures. Our review addresses this by providing
deep analysis targeting health data, including but not
limited to medical records, ECG and genomic data.
We examine re-identification methods and assess
standard practices, techniques and factors leading to
successful breaches to inform the debate on balancing
privacy and utility. Our investigation introduces cat-
egorizations of de-anonymization attacks specific to
health data and suggests a novel framework for eval-
uating anonymization methods. This framework con-
siders the unique characteristics of health datasets and
advanced analytics, including machine learning (ML)
and deep learning (DL), assessing the availability of
open-access de-anonymization codes and models and
reaching out for access, highlighting the importance
of reproducibility. Through this critical analysis, we
identify significant gaps and pinpoint suggestions for
innovative directions.
Aguelal, H. and Palmieri, P.
De-Anonymization of Health Data: A Survey of Practical Attacks, Vulnerabilities and Challenges.
DOI: 10.5220/0013274200003899
In Proceedings of the 11th International Conference on Information Systems Security and Privacy (ICISSP 2025) - Volume 2, pages 595-606
ISBN: 978-989-758-735-1; ISSN: 2184-4356
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
595

1.1 Contributions and Novelty
Through this survey, we introduce a new framework
and identify gaps. The key contributions are:
• Novel Attack Categorization Framework: In-
troducing a new taxonomy and framework to as-
sess de-anonymization attacks on health data:
1. Dataset types and sources.
2. Methodologies employed with a review of
available or shared models.
3. Success rate evaluation and comparing them to
understand effectiveness better.
4. Incorporation of advanced techniques by anal-
ysis of recent ML and DL methods applied to
de-anonymization missing in previous surveys.
• Practical Emphasis: Unlike existing reviews fo-
cusing on hypothetical risks, we offer insights into
actual risks on practical, real-world attacks, bridg-
ing the gap between research and practice.
• Assessment of Reproducibility, Generality and
Code Availability: Evaluating the availability of
codes to underscore the challenges in reproducing
studies and the importance of open-source prac-
tices and identify gaps in attack generality.
• Ethical and Legal Considerations: We look
at the ethical and legal guidelines, focusing on
GDPR and HIPAA as standards.
• Key Recommendation: Based on the results and
challenges, we suggest recommendations such as
establishing standardized benchmarks for evaluat-
ing de-anonymization in healthcare.
This paper’s remaining sections are arranged as
follows: Sec. 2 covers the background on vulnera-
bilities in health datasets and related works. Sec. 3
outlines the methodology, including selection criteria
and data extraction. Sec. 4 reviews de-anonymization
techniques, categorization, and evaluation. Sec. 5
presents key insights and findings, while Sec. 6 dis-
cusses strengths, gaps, and recommendations. Fi-
nally, Sec. 7 summarizes contributions and future re-
search directions.
2 BACKGROUND AND RELATED
WORK
De-anonymization (re-identification) links back or
extracts anonymized data to individuals, posing a
unique privacy threat, especially with the uniqueness
of medical data. Despite the use of k-anonymity
(Sweeney, 2002), l-diversity (Machanavajjhala et al.,
2007), and t-closeness (Li et al., 2006) (Bayardo and
Agrawal, 2005), vulnerabilities persist and require
practical assessment. Adversaries exploit patterns,
unique identifiers Table 1, and correlations to un-
dermine traditional anonymization. The rise in pub-
lic datasets post-2009 (Henriksen-Bulmer and Jeary,
2016) eased access to auxiliary information, driving
attacks. While regulations emphasize anonymization,
we note challenges, especially in healthcare, requiring
approaches. This motivates our systematic evaluation
of de-anonymization effectiveness and field insights.
2.1 Related Work
Several studies have explored data de-anonymization
techniques across various domains. (Ding et al., 2010;
Bhattacharya et al., 2023; Ji et al., 2019) focus on
social networks, utilizing graph-based structures and
interactions. Similarly, (Farzanehfar et al., 2021)
examined geolocation by tracing individuals’ move-
ments. Surveys such as (Al-Azizy et al., 2016) and
(Henriksen-Bulmer and Jeary, 2016) reviewed gen-
eral de-anonymization methods, including link pre-
diction and data aggregation. However, they lacked
a focus on health data or practical feasibility.
In healthcare, early works (Sweeney, 1997)
demonstrated the re-identification of medical records
by linkage to publicly available voter registration
data, and (Malin and Sweeney, 2004) on genomics
using trail re-identification techniques. Authors in
(Prada et al., 2011) summarize the regulatory efforts
and overview of risks in healthcare, and in (Emam
et al., 2011), researchers reviewed anonymization
techniques and limitations, particularly with smaller
datasets. However, their work is dated and does
not reflect on emerging empirical issues and practi-
cal de-anonymzaition of health datasets. Earlier re-
views did not fully account for recent advancements
in ML/DL that have changed de-anonymization capa-
bilities, consequently, the need for our survey. For
instance, (Shokri et al., 2017) introduces his mem-
bership inference attack (MIA) against ML models
to show how adversaries can determine if a specific
record was part of the model’s data (Nasr et al., 2019).
(Lee et al., 2017) presented blind attack using gener-
ative adversarial networks (GANs) and DL in bypass-
ing anonymization, (Yin et al., 2023) and (Lu et al.,
2024) emphasize the increasing threat posed by so-
phisticated DL algorithms.
2.2 Legal and Ethical Considerations
De-anonymization of health data raises legal and eth-
ical concerns. The need to protect Personally Iden-
ICISSP 2025 - 11th International Conference on Information Systems Security and Privacy
596
tifiable Information (PII) and Quasi-Identifiers (QIs)
is necessary; regulations emphasize removing or gen-
eralizing identifiers (see Table 1) and according to
GDPR (European Union, 2016): ” Personal data
means any information relating to an identified or
identifiable ... by reference to an identifier such as
name, an identification number, ... or factors specific
to the physical, physiological, genetic...”
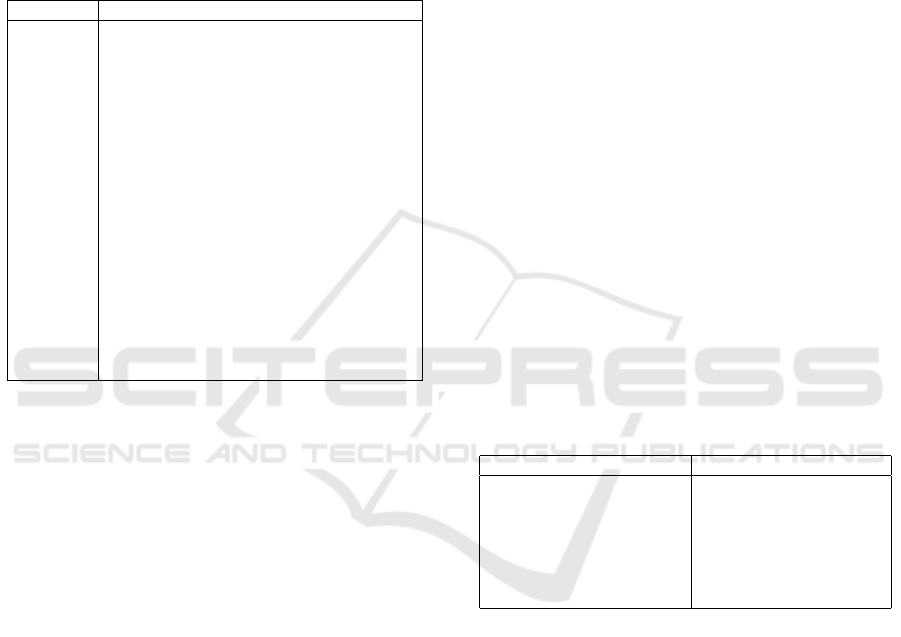
Table 1: The 18 elements in the HIPAA must be removed
or generalized for a data set to be de-identified.
Identifier Description
(A) Names
(B) Geographic subdivisions smaller than a State
(C) All elements of dates
(D) Telephone numbers
(E) Fax numbers
(F) Electronic mail addresses
(G) Social security numbers
(H) Medical record numbers
(I) Health plan beneficiary numbers
(J) Account numbers
(K) Certificate/license numbers
(L) Vehicle identifiers and serial numbers
(M) Device identifiers and serial numbers
(N) Web URLs
(O) IP address numbers
(P) Biometric identifiers
(Q) Full face photographic images
(R) Any other unique identifying number, char-
acteristic, or code
However, de-anonymization can lead to potential
breaches and ethical implications, among others:
• Patient Trust: Re-identification weakens trust be-
tween patients (BEUC, 2023) and providers.
• Informed Consent: Patients must be fully aware
of the risks associated with data sharing.
• Data Utility vs. Privacy: Balancing the utility of
health data against the need to protect individual
privacy is a persistent dilemma.
Our review considers legal and ethical dimensions
and accurately assesses de-anonymization to improve
data privacy in the health sector.
3 METHODOLOGY
This section outlines our approach, including a de-
fined scope, the selection process, inclusion/exclusion
criteria, and the data extraction process. We followed
rigorous guidlines adopted from the established pro-
cedures (Kitchenham, 2004) to ensure the empirical
relevance of the reviewed studies.
3.1 Scope of this Review
This review focuses exclusively on de-anonymization
attacks targeting health-related information. It en-
compasses a range of health data types, including
medical records, neuroimaging data, ECGs, wearable
device data, and genomic data. We emphasize arti-
cles that discuss practical or simulations, providing
empirical evidence on health data rather than theoret-
ical analysis to address the practical risks.
The initial search returned 1170 papers, narrowed
to 146 after initial screening, with 69 deemed rele-
vant following abstract analysis. We ended up with
17 papers in the summary with empirical evidence
on health data types. We explored the availability of
codes, and only five works explicitly provided access
with one inaccessible link. For the rest, we contacted
the authors directly, and two of them provided access.
This highlights the varying levels of transparency and
accessibility regarding reproducibility.
3.2 Search Strategy
We constructed a detailed search strategy utilizing
Boolean operators and a set of the related keywords
combination Table 2. We supplemented these with
terms like ”Data Privacy Risk” and ”Identification”
to ensure comprehensive coverage.
Table 2: Search terms.
Primary search terms Some excluded terms
De-anonymizaiton Social Networks
Re-identification Vehicle data
Deanonymization attacks Smart city data
Anonymizaiton Assess-
ment
Marketing and Finance
Analytics
Identification Location and Geolocation
Health data
We performed our initial search on the listed
databases to capture literature from fields: health in-
formatics, computer science, and privacy studies:
• IEEE Xplore
• ACM Digital Library
• Google Scholar
• PubMed
• SpringerLink
Additionally, we manually reviewed references to
avoid missing relevant work.
3.3 Inclusion and Exclusion Criteria
We developed our inclusion criteria, Table 3, to meet
our objectives and research questions.
De-Anonymization of Health Data: A Survey of Practical Attacks, Vulnerabilities and Challenges
597
• What data types and corresponding de-
anonymization studies have been most targeted?
• Did the authors perform or develop an attempt
of de-anonymization attack? What types of at-
tributes, were used?
• What datasets were used in these de-
anonymization studies, and how they char-
acterized in size and diversity?
• How to interpret the results and outputs presented
in the works?
• To what extent the studies address the repro-
ducibility and transparency, such as providing ac-
cess to code?
Table 3: Inclusion Criteria for Study Selection.
Criterion Description
Data Types Involve health data types, including
but not limited to genomic data, ECG
and medical records.
Study Type &
Empirical Vali-
dation
Studies detail empirical attempts,
practical or simulations.
Results and
Quality of
Methodology
Studies must present quantifiable re-
sults, including success rates and de-
tailed methodologies.
Peer-Reviewed
Publications
Studies in peer-reviewed journals or
conference proceedings.
Language English-language studies for consis-
tent analysis and interpretation.
Publication Date Studies published from 2010 onwards
Exclusion Criteria:
1. Duplicate or Redundant Publications.
2. Non-Health Data Focus: We excluded studies
outside the healthcare context.
3. Theoretical Studies: We excluded works focus-
ing solely on theoretical analyses.
4. Lack of Methodological and Result Detail:
Studies with insufficient experimental validation
and measurable results.
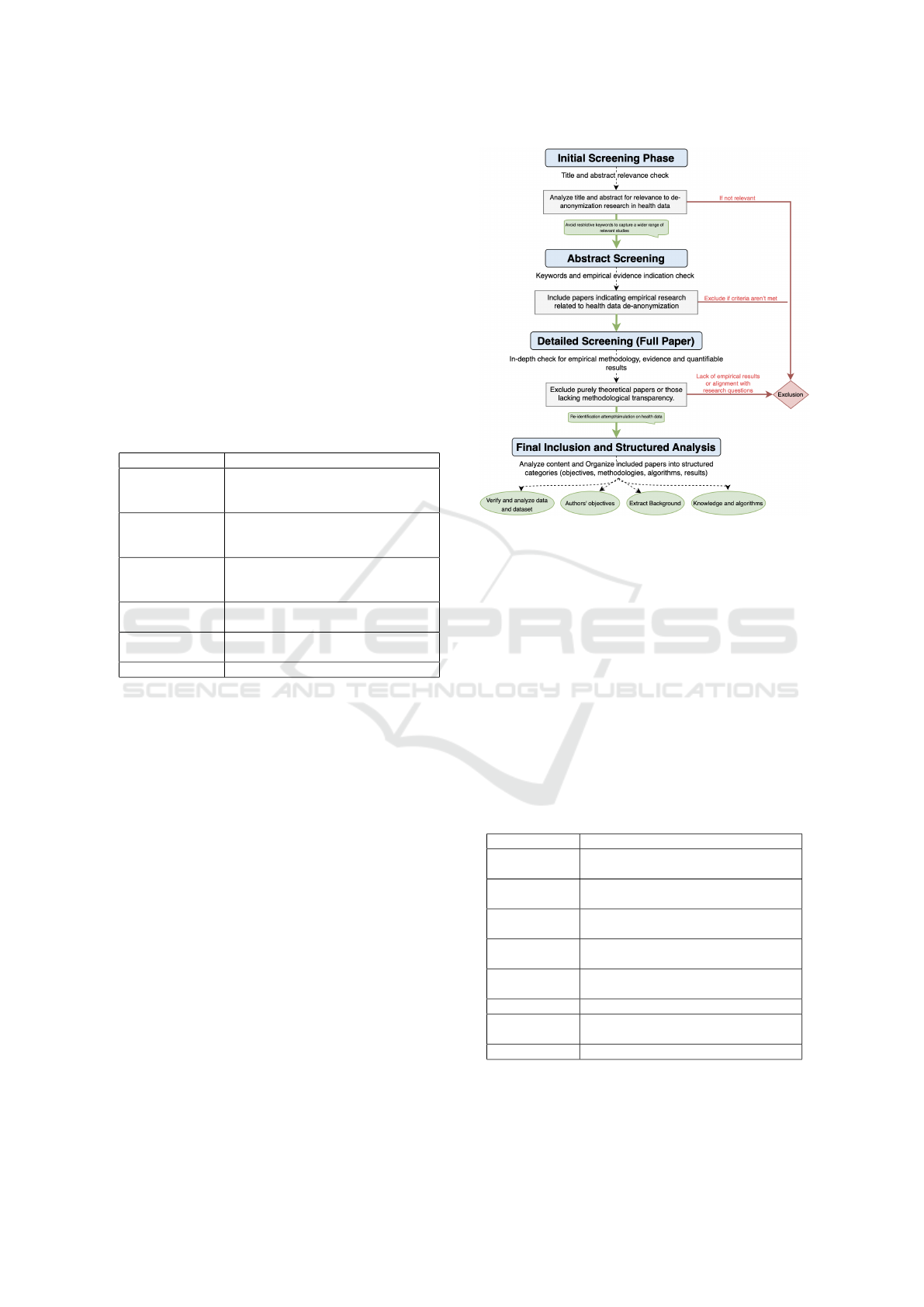
3.4 Data Extraction Process
We followed a structured data extraction process
(Fig. 1) with four phases:
1. Identification: We initially reviewed the studies’
titles, abstracts, and keywords to assess relevance.
2. Abstract Screening: Detailed abstract analysis.
3. Core Eligibility: Full-text review to match re-
search questions.
4. Final Inclusion: Studies offering insights into at-
tacks’ applicability and methodologies.
Figure 1: Data extraction and selection process.
Through this multi-phase process, we narrowed
the selection to 17 papers that provide empirical ev-
idence attacks and measurable outcomes. We then
further extracted insights, methodologies and success
rates aligning with our scope, inclusion 3 and exclu-
sion criteria.
3.5 Structured Analysis
We organized the extracted information into a struc-
tured format to classify and evaluate the findings’ crit-
ical components, as shown in Table 4.
Table 4: Structured Analysis of De-Anonymization Re-
search in Health Data.
Section Information
Title Analysis of the title and implications
for de-anonymization in health data.
Abstract Summary of aims, methodology, re-
sults, and significance.
Methodology
& Objectives
Detailed approach and research goals.
Data Source
& Data type
Inventory of data used and sources.
Background
& Algorithm
Theoretical foundation and algorithms
used.
Attack Type Classification of the attack(s) studied.
Results & Ob-
servations
Key findings success rates and in-
sights.
Limitations Constraints and future research areas.
ICISSP 2025 - 11th International Conference on Information Systems Security and Privacy
598
4 SURVEY OF
DE-ANONYMIZATION
TECHNIQUES
This section surveys the techniques applied to health
information. We split the processes and assessed the
used approaches. Our deconstruction forms a frame-
work based on categories, data types, and methods
and describes strategies and findings.
4.1 Data Types and Analysis
We categorize attacks by data types vulnerable to de-
anonymization and detailed vulnerabilities in Table 5.
1. Genomic Data: Genomic data is a key target
due to its unique nature. Studies such as (Gym-
rek et al., 2013; Humbert et al., 2015; Lippert
et al., 2017; Wan et al., 2021) exploit single-
nucleotide polymorphisms (SNPs) and open ge-
netic databases for re-identification. Additionally,
(Ayoz et al., 2021; Thenen et al., 2019) use partial
genetic data based on data beacons.
2. Medical Records (Including EHRs): Medical
records often contain sensitive information, par-
ticularly quasi-identifiers (QIs). (Ji et al., 2020;
Branson et al., 2020) explored linkage and infer-
ence attacks on EHRs using probabilistic models.
Furthermore, (Antoniou et al., 2022) emphasized
the risk associated with k-anonymity, highlighting
the need for stronger anonymization.
3. Neuroimaging Data: Neuroimaging data, such
as MRI scans, contain unique patterns in brain
structures discussed in (Ravindra and Grama,
2021; Venkat-esaramani et al., 2021) to exploit
deep learning (DL), notably convolutional neural
networks (CNNs), in identifying individuals.
4. Electrocardiograms (ECGs): Due to their
uniqueness, ECG signals have been increasingly
used for re-identification (Ghazarian et al., 2022).
Studies like (Min-Gu Kim, 2020), (Hong et al.,
2020), and (Mitchell et al., 2023) demonstrate
various ECG-based re-identification techniques
and implement models such as Support Vector
Machines (SVM), highlighting vulnerabilities ex-
ploited for attacks.
5. Wearable Device Data: Wearable devices gen-
erate continuous streams of health-related data,
such as vital signs. (Lange et al., 2023) and (Min-
Gu Kim, 2020) demonstrated that similarity-
based attacks, founded on techniques like Dy-
namic Time Warping (DTW), can align time-
series data and re-identify users based on vital pat-
terns.
In Sec.5 we further explore the deconstruction and
key insights (Table 6, Table 7).
4.2 Classification of Attacks
To address the concerns about the attack classifica-
tion, and based on adversaries’ objectives and tech-
niques, we organize them into consistent categories to
address gaps and overlaps in the literature and ensure
each class is distinct and self-contained. While these
are known attacks applied in various contexts, cur-
rent classifications are often inconsistent, with some
classes overlooked or ambiguously grouped. Our
framework aims to clarify these distinctions and pro-
vide a reference for future research.
While a unified threat model is not explicitly de-
fined, we categorize de-anonymization under estab-
lished criteria. Each study inherently defines its own
threat model tailored to the dataset, methodology,
and objectives. By evaluating each attack, we indi-
rectly address the adversarial settings relevant to these
works.
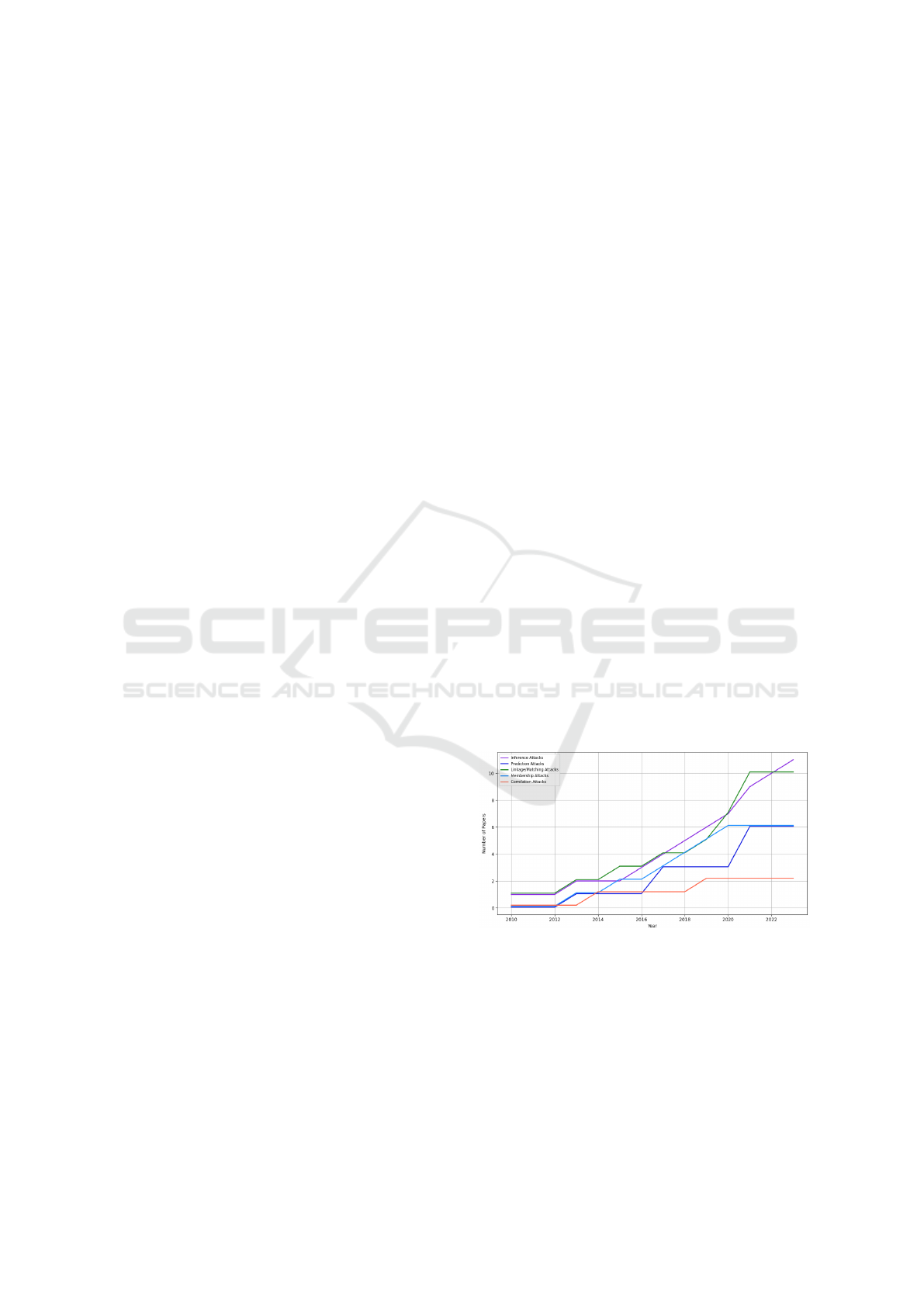
4.2.1 Inference Attacks
Settling as one of the most common classes (see
Fig. 2), involves extracting sensitive information us-
ing ML/DL (Wu et al., 2020) and aggregating hidden
patterns from the same or multiple sources. Discussed
in (Shokri et al., 2017; Thenen et al., 2019) by deduc-
ing attributes and use of auxiliary information.
Figure 2: Evolution of De-anonymization Techniques Over
Time (Health Data).
4.2.2 Linkage or Matching Attacks
Linkage or matching attacks connect records from
different datasets to identify individuals using aux-
iliary information, as demonstrated in (Venkat-
esaramani et al., 2021). These frequent attacks form
a foundational class of de-anonymization methods.
De-Anonymization of Health Data: A Survey of Practical Attacks, Vulnerabilities and Challenges
599
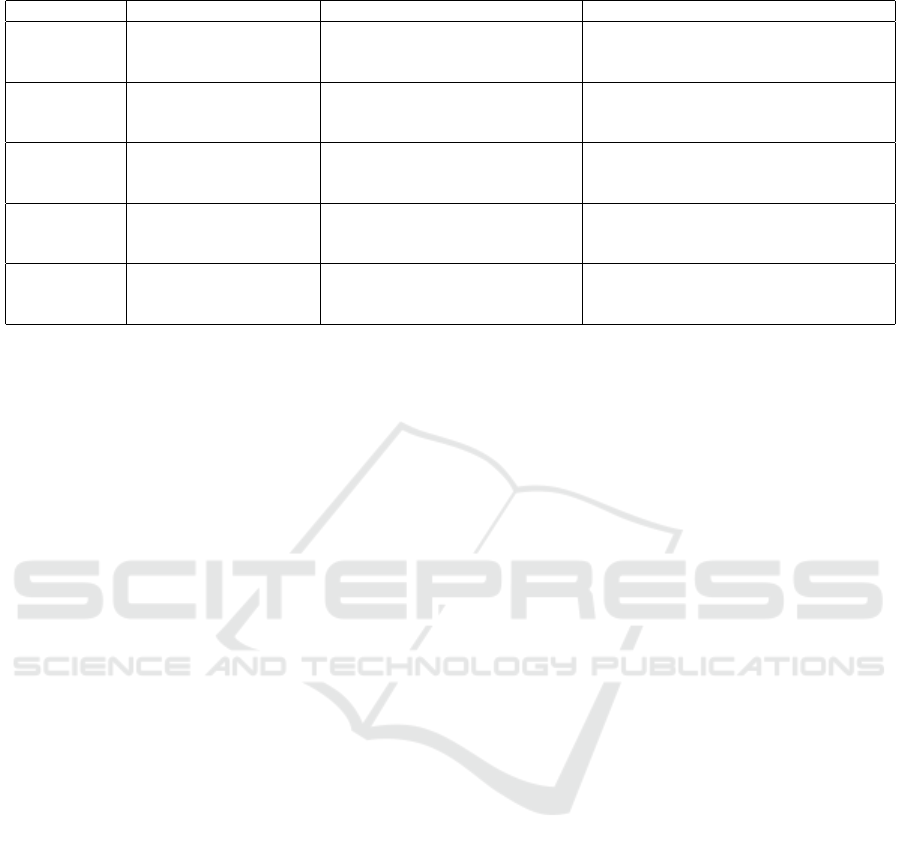
Table 5: De-Anonymization Vulnerabilities and Techniques by Health Data Type.
Data Type Key Vulnerabilities Techniques Used in Attacks ML/DL Methods and Modifications
Genomic
Data
Unique genetic markers
(e.g., SNPs), open ge-
nealogy dbs
SNP linkage; allele frequency
comparisons; mutation patterns
and beacons exploitation
Feature selection for SNP density, RNNs
for sequential patterns; Probabilistic and
regularization for diversity
Medical
Records and
EHR
Sensitive QIs; health
conditions and at-
tributes inferences
Linkage with auxiliary data;
probabilistic models, stylometric
analysis and Bayesian models
CNNs with attribute encoding layers,
QIs feature correlation analysis; RNNs
for temporal sequence data
Neuroimaging
(e.g., MRI)
Unique patterns in
brain structures, high-
dimensional features
Deep feature extraction using
CNNs, structural matching and
spatial features capture
CNNs and transfer learning to leverage
pre-trained models; optimized layers to
isolate unique brain patterns
ECG Data Heartbeat variability,
signal characteristics,
periodic waveforms
Signal decomposition; DTW and
fiducial/non-fiducial methods for
feature and temporal variations
LSTM for time dependencies; hybrid
CNN-RNN, regularization and data aug-
mentation to increase accuracy
Wearable
Device Data
Continuous metrics,
lifestyle identifiers,
vital signs
DTW; cross-user sensor patterns’
and behaviour-based tracing
Multi-modal CNNs for sensor fusion
data, clustering for temporal similarity;
attention layers for pattern recognition
4.2.3 Membership Attacks
Designed to determine if an individual’s data is en-
closed in a dataset, which challenges anonymization
by demonstrating that individuals can be pinpointed.
The concept of MIA has been tested and validated
(Ayoz et al., 2021)and discussed in the literature as
a threat to the privacy of health datasets, even with
the current privacy-preserving solutions as synthetic
data generation (Zhang et al., 2022).
4.2.4 Prediction Attacks
Prediction attacks, though related to inference attacks,
focus precisely on predicting sensitive attributes or
traits. This attack aims to forecast characteristics such
as phenotypic traits in our scope, (Lippert et al., 2017)
demonstrated its effectiveness in genomics. These at-
tacks are distinct from inference ones due to the focus
on forecasting instead of aggregation.
4.2.5 Correlation Attacks
Exploit the statistical relationships between
anonymized and external data as (Ji et al., 2020)
defined and simulated similarly to inference, but
the suggestion is to distinguish this class for the
reliance on cross-referencing statistical relationships.
(Narayanan and Shmatikov, 2008) highlight the
power of correlation and validate this as a strategy.
4.2.6 Justification of Classification
Our classification is founded on the literature and ap-
plications in different domains. The distinctions are
based on the adversary’s objectives and the specific
techniques. Some papers merge certain types for the
overlapping methodologies, but we chose to separate
them to reflect the differences acknowledged.
4.3 Methodologies Summary
This section reviews the diverse methodologies and
techniques extracted from ML/DL techniques to
probabilistic models. Below, we summarise the meth-
ods and principles for piloting the approaches.
1. Machine Learning and Deep Learning
ML and DL are fundamental in health, enabling
de-anonymization through feature extraction from
complex datasets. DL was used to identify unique
patterns (Wu et al., 2020), CNNs achieved over
94% accuracy in neuroimaging (Ravindra and
Grama, 2021), and unsupervised learning linked
facial and genomic data (Venkat-esaramani et al.,
2021). The deployment of ML/DL leverages cor-
relations to bypass traditional anonymization, as
RNNs (e.g., LSTM) for ECG rhythm variations’
identification and waveform morphologies, CNNs
to capture neuroimaging’s spatial hierarchies in a
pixel pattern and transformers on features across
sequences like in SNP or EHR with distant rela-
tionships between data points.
2. Probabilistic and Statistical Learning Models
Probabilistic models used for genomic data and
estimating phenotypic traits, (Humbert et al.,
2015) combined unsupervised and supervised
learning to predict phenotypic traits and map them
to identities (Gymrek et al., 2013), and (Shringar-
pure and Bustamante, 2015) employed Bayesian
inference and hypothesis testing to detect genome
presence, but we highlight the effectiveness’s de-
crease in larger populations.
3. Clustering and Similarity-Based Techniques
These methods exploit inherent patterns and out-
liers in time series and wearable data. (Lange
et al., 2023) used DTW to achieve a 70.6%
re-identification rate on the WESAD dataset
ICISSP 2025 - 11th International Conference on Information Systems Security and Privacy
600
(Schmidt et al., 2018). K-NN, and similar al-
gorithms analyze distinctive behavioral patterns,
such as ECG peaks and fiducial biometrics (Min-
Gu Kim, 2020; Hong et al., 2020).
4. Stylometric and Correlation Techniques
Used mainly to analyze text-based EHR or similar
data by exploiting unique writing styles. (Ji et al.,
2020) presented stylometric techniques to match
data using online profiles, which also exploit ex-
ternal metadata (Saxena et al., 2024).
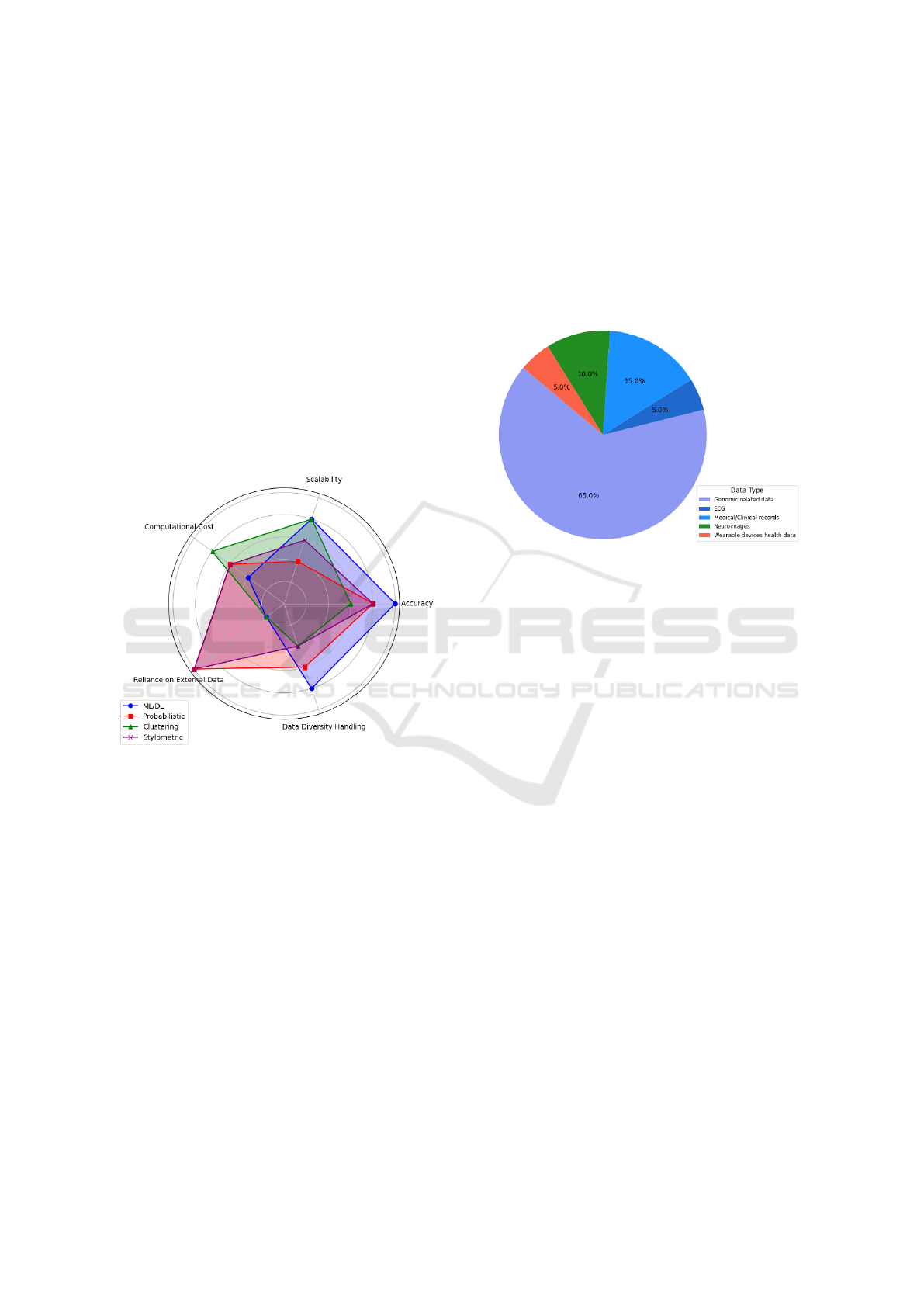
Each methodology was evaluated on a scale from
1 to 5 on five key attributes (Accuracy, Scalability,
Computational Cost, Reliance on External Data, Data
Diversity Handling) to underscore the strengths and
weaknesses in its applicability to de-anonymization.
The chart in Fig. 3 provides a summary illustrating
how each method performs.
Figure 3: Comparison of Methodologies based on 5 Key
Attributes.
5 FINDINGS
This section highlights key findings from the sur-
veyed practical studies. Table 6 summarizes attacks
on health datasets based on the evaluated criteria.
5.1 Key Insights
From our analysis of methodologies and findings, we
highlight several key insights:
• Data-Specific Focus: Attacks exploit specific
characteristics and vulnerabilities (see Table 5).
1. Genomic Data Vulnerability: Ranked as the
most targeted due to its uniqueness, Fig. 4.
(a) SNP and genetic genealogy use demonstrated
success rates >80% in (Shringar-pure and
Bustamante, 2015; Edge and Coop, 2020).
(b) Beacons and genome reconstruction (Ayoz
et al., 2021) for information inference.
2. Neuroimaging Data: Exploits unique brain
patterns in MRI scans.
3. ECG and Wearable Device Data: Vulnerable
to attacks using temporal patterns.
Figure 4: Distribution of Data Types Trageted.
• Success Rate Trends: Success rates vary depend-
ing on the strategy Fig. 5, and commonly decrease
as dataset size increases. We explain it with:
1. Increased Data Diversity: Larger datasets in-
troduce variability (Gymrek et al., 2013), con-
fusing the identification of unique patterns,
2. Noise in the Data: Increased noise blurs
unique markers.
3. Scalability: ML/DL models can be effective
but struggle with heterogeneous datasets and
require high computational resources.
• Dependency on Auxiliary Information: Attack
success often relies on auxiliary data, such as de-
mographic data and beacons for genomics as in
(Thenen et al., 2019; Ayoz et al., 2021).
• Variation on Explainability: Probabilistic and
clustering methods offer greater clarity (Venkat-
esaramani et al., 2021), while some studies lack
detail due to their methodologies.
• Uniqueness and Sensitivity: Health data
(e.g., biological patterns) requiring specific re-
identification and analysis techniques.
• Dominance of Certain Attacks: Inference and
membership attacks are predominant due to the
adaptability (see Fig. 2), unlike correlation that
depends on the auxiliary data availability.
• Ethical Considerations: Studies followed ethi-
cal guidelines, balancing research progress with
De-Anonymization of Health Data: A Survey of Practical Attacks, Vulnerabilities and Challenges
601

Table 6: Summary of De-Anonymization Vulnerabilities, Techniques and Results by Health Data Type.
Work Main Studied
Data
Datasource Background Knowl-
edge and Technique
Attack Strat-
egy
Results
(Min-
Gu Kim,
2020)
ECG MIT-BIH
NSRDB
Noise removal, 2D
ECG image transfor-
mation, CNN, RNN
Inference at-
tacks
98.9% recogni-
tion (ensemble
network)
(Lange et al.,
2023)
BVP, EDA,
TEMP, ACC
WESAD dataset DTW alignment algo-
rithms, similarity rank-
ing
Similarity-
based inference
attack
70.6% identifica-
tion at k=1
(Venkat-
esaramani
et al., 2021)
3D face im-
ages, SNPs
OpenSNP, LFW,
MegaFace,
CelebA
DL (CNN, VGGFace),
face-to-phenotype pre-
diction
Prediction,
matching at-
tack
80% for small
populations,
<20% for popu-
lations >100
(Humbert
et al., 2015)
SNPs, pheno-
types
OpenSNP,
SNPedia,
23andMe,
FTDNA
Supervised ML, proba-
bilistic models
Inference,
matching at-
tacks
52% success (su-
pervised) for 10
participants
(Gymrek
et al., 2013)
Genetic geneal-
ogy (Y-STR)
Ysearch, inter-
net research
Illumina sequencing,
lobSTR algorithm
Inference, link-
age attacks
12% surname re-
covery
(Ravindra
and Grama,
2021)
Neuroimaging
data
HCP, ADHD-
200
Leverage-score sam-
pling, clustering-based
approach
Matching, in-
ference attacks
94% accuracy for
HCP; 97.2% for
ADHD
(Edge and
Coop, 2020)
Genetic geneal-
ogy
GEDmatch IBS tiling, probing,
baiting
Inference, link-
age attacks
82% genome re-
covery with IBS
Tiling
(Ji et al.,
2020)
Medical
records
WebMD,
HealthBoards
Stylometric, correlation
features, SVM, KNN
Correlation at-
tacks
12.4% re-
identification
(Wu et al.,
2020)
Medical
records, mam-
mography
images
Cardiovascular
Disease Dataset,
MIAS, CBIS-
DDSM
Attribute inference,
model inversion, CNN,
MLP
Inference at-
tack
80% attack suc-
cess without de-
fenses; defenses
reduced accuracy
(Branson
et al., 2020)
Medical reports Nepafenac, U.S
Public death
and discharge
Records, FOIA,
Facebook, Red-
dit
Manual pattern match-
ing, auxiliary informa-
tion correlating and
record linkage
Inference, link-
age attacks
6 low-confidence
matches out of
500 patients (pro-
cess consumed
170 hours)
(Thenen
et al., 2019)
Genomic data
(SNPs)
HapMap QI-attack (LD cor-
relations), GI-attack
(Markov chain)
Inference at-
tacks
282 queries for
95% confidence
(Shringar-
pure and
Bustamante,
2015)
Genomic data
(beacons)
1000 Genomes Likelihood-ratio test
(LRT)
Membership at-
tack
>95% detection
with 5,000 SNP
queries
(Ayoz et al.,
2021)
Genomic data
(beacons)
OpenSNP,
HapMap
Clustering, SMOTE,
ensemble classifiers
Prediction,
membership,
inference at-
tacks
0.96 precision for
SNP reconstruc-
tion
(Lippert
et al., 2017)
Genomic data 1,061 individ-
uals from San
Diego
PCA for face variation,
ridge regression, max
entropy models
Prediction,
linkage attacks
80% in mixed co-
hort, 50% in sub-
cohorts
(Ayday and
Humbert,
2017)
Genomic data CEPH/Utah
Pedigree 1463,
1000 Genomes
Project
Belief propagation,
graphical models,
Markov models
Inference,
membership
attacks
87% in inferring
hidden SNPs from
multiple relatives,
(Wan et al.,
2021)
Genomic data Craig Venter’s
record, Ysearch,
Publicfinders
Backward induction,
greedy algorithm
Inference, link-
age attacks
76% in no-
protection sce-
nario
(Liu et al.,
2018)
Genetic se-
quencing,
phenotype data
eMERGE-PGx,
VUMC SD,
KPW, NW
Likelihood ratio, mem-
bership detection
Membership at-
tacks
80% re-
call/precision
for 16,346 indi-
viduals
ICISSP 2025 - 11th International Conference on Information Systems Security and Privacy
602
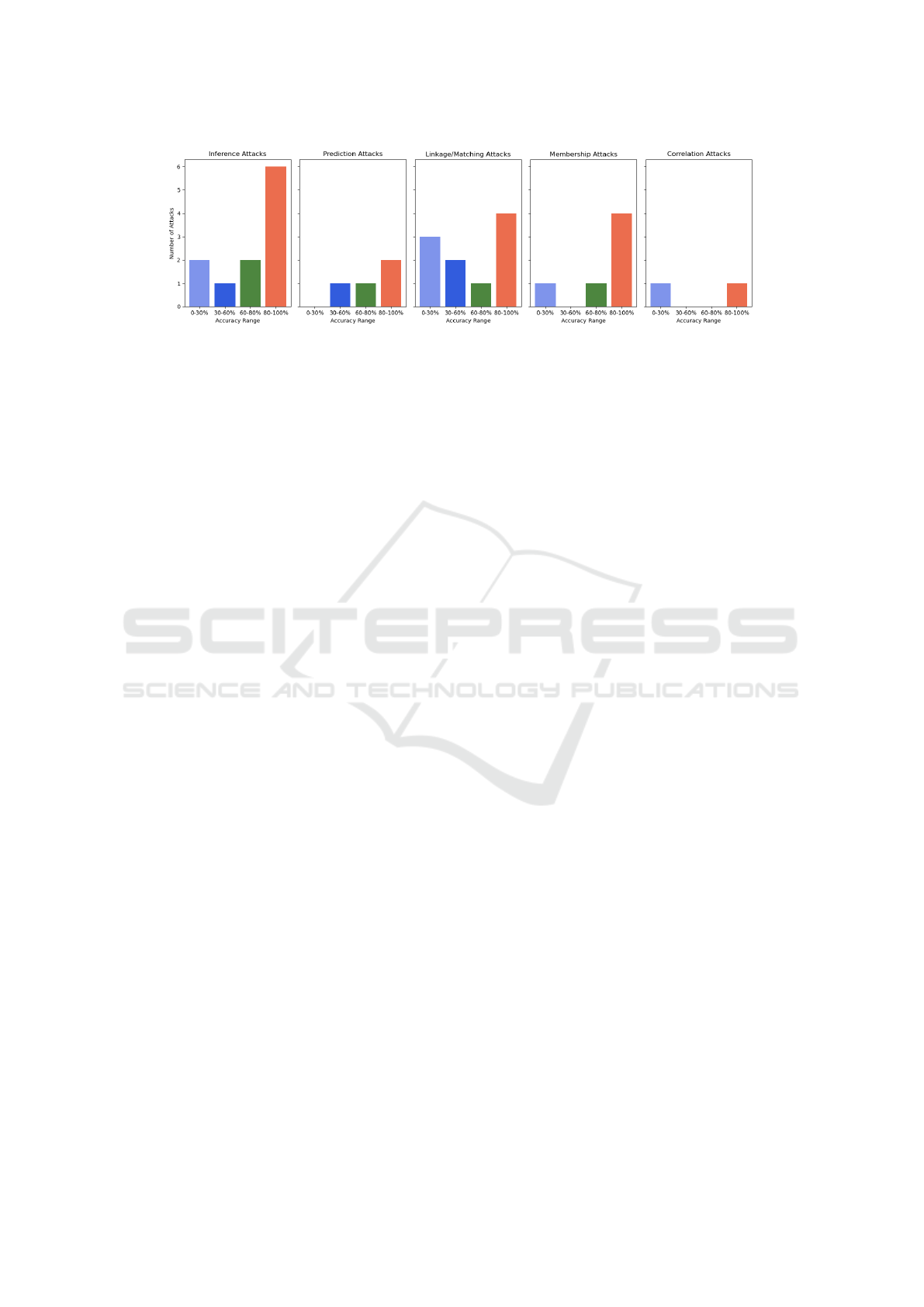
Figure 5: Ranges of Success Rates of De-anonymization Attacks.
privacy. However, de-anonymization remains a
threat, requiring regulatory updates.
Table 7 shows a comparative analysis and assess-
ment based on effectiveness and common vulnerabil-
ities and completes the breakdown done in Sec.4.
6 EVALUATION AND
DISCUSSION
We deliver a critical lens on the literature’s strengths
and challenges, through which we can observe de-
anonymization with rational considerations.
6.1 Key Strengths
• High Accuracy Rates: Reporting high success
when using multiple sources. See Fig. 5.
• Verification of Hypotheses: The simulation on
real-world datasets (Connectome and 23andMe)
indicates practical relevance and potential impact.
• Diverse Methodologies: Various methodologies,
including CNNs, RNNs, clustering and proba-
bilistic models, define the multiple attack types.
• Innovative Attack Techniques: DL and stylo-
metric and behavioural analysis (people’s interac-
tion with data), as (Ji et al., 2020) debate the evo-
lution of methods beyond standard approaches.
• Adaptability to Evolving Data: Flexible to
evolving technologies and emerging data formats
(e.g., IoT-related)(Dimitrievski et al., 2023).
6.2 Challenges
Our work reveals challenges to address in anonymity
assessment. We delve more into these gaps:
• Reproducibility Challenge: The absence of pub-
licly available code, use of restricted or non-
public datasets (e.g., (Lippert et al., 2017; Bran-
son et al., 2020)) and methodological descrip-
tions insufficience restrains verification, testing
and hinders reproducibility.
• Scalability of Studies: While some studies use
large datasets (Thenen et al., 2019), other works
were not tested broadly (Min-Gu Kim, 2020) rais-
ing scalability and practicality concerns.
• Underreporting of Negative Results: We high-
light a direction to underreport negative results in
de-anonymization. By not publishing unsuccess-
ful attempts, we risk losing valuable insights.
• Under-Representing Data Types: Several health
data types were studied. However, other valuable
data have not been explored in de-anonymziation,
such as pathology images and speech data.
• Interdisciplinary Approaches & Application
Scope: Current research often lacks integration
across different disciplines and broader applica-
tions, such as fraud detection.
• Lack of Assessment on Advanced Anonymiza-
tion Techniques: We debate the lack of applica-
ble attempts on emerging anonymization namely
Differential Privacy (DP) and advanced synthetic
data generation, holding back our understanding
of the robustness of these methods in real world.
• Poor Explainability: The interpretability of the
white-box models remains limited. Explainable
models will improve transparency in assessments.
• Ambiguity in Legal Regulations: GDPR and
HIPAA lack precise criteria for what is suffi-
ciently anonymized data. An apprehension in
clear legal guidance on de-anonymization risks
can slow efforts to assess patients’ privacy.
6.3 Recommendations
Building upon our survey and the identified gaps, we
suggest recommendations:
1. Establishement of Standardized Benchmark-
ing and Datasets.
De-Anonymization of Health Data: A Survey of Practical Attacks, Vulnerabilities and Challenges
603

Table 7: Insights and Analysis of De-Anonymization Techniques.
Category Insights Techniques
Comparative Suc-
cess Rates
- Higher success in genomic & neuroimaging data
- Text-based & wearable data require auxiliary info for accuracy
Genomic & Neuro: ML/DL
models; Wearable: Clustering
Methodological
Performance
- ML/DL: Best for structured data (e.g., imaging); CNNs for spa-
tial data, RNNs/LSTMs for time dependencies
- Probabilistic: Effective for probability-based genomic matches,
- Clustering/Similarity: Ideal for repetitive time-series patterns
CNNs, RNNs, LSTM, Bayesian
Inference, likelihood-ratio test-
ing, DTW
Common Vulnera-
bilities
- Genetic markers, biometric patterns (e.g., ECG)
- Auxiliary data (genealogy records, beacons) for linkage
ML/DL, Clustering, Similarity
and Probabilistic models
Feature Extraction - CNNs & DL improve re-identification through feature isolation
- Clustering & DTW scalable but need quality data
DL models for high accuracy
Evaluation & Scal-
ability
- CNNs: High accuracy, lower efficiency
- Clustering: Scalable, less accurate in noisy environment
CNNs vs Clustering for varied
data adaptability
We suggest developing standardized datasets and
benchmarks for evaluating de-anonymization in
health to facilitate accurate results comparisons
and reproducibility and foster collaboration. Our
advocacy for common benchmarks, including
health data types and anonymization levels, can
include:
• Organized datasets with variables for testing.
• Standard metrics like success rates, accuracy,
and computational efficiency.
• Open shared repositories.
2. Promote Reproducibility and Open Science
Practices.
We recommend publicly sharing code, data, and
methods to improve reproducibility, result valida-
tion and study replication. Key steps include:
• Using open-source licenses and documentation.
• Use accessible repositories.
• Complies sharing with ethics and regulations.
3. Enhance Scalability Testing and Evaluation.
Assess attacks on large, diverse datasets to reflect
real-world constraints. Our success rate evalua-
tion (Fig. 5) highlights the need for scalability.
4. Encourage Publication of Negative Results.
We advocate for a shift in the research commu-
nity to overcome the under-reporting of negative
results. Sharing negative findings provides valu-
able insights, mainly preventing redundant efforts
to achieve more reporting transparency.
5. Expand Research to Wider Health Data Types.
We recommend more interdisciplinary collabora-
tion in computer science, data privacy, health-
care and bioinformatics; to open new opportuni-
ties exploring underrepresented types like voice
and speech, pathology images and dietary data.
6. Pratical Assessment of Advanced Anonymiza-
tion Techniques.
We suggest conducting practical attacks on meth-
ods like DP and synthetic data generation in
health. We promote publishing successful and un-
successful results.
7. Broaden the De-Anonymizaiton Applications.
We advocate to explore broader applications
of de-anonymization techniques beyond re-
identification, such as:
• Fraud Detection: Using de-anonymization
(e.g., pattern recognition, cross-referencing) to
identify fraudulent activities in healthcare.
• Data Integrity Verification: Anonymized data
tamper detection and provenance tracking.
While our paper evaluates de-anonymization tech-
niques and challenges, an extended version will elab-
orate on comparative evaluation, validation, and the
detailed benchmarking framework.
7 CONCLUSION
Our survey underscores the state of de-anonymization
in healthcare, given the metrics, approaches, and data
features used in different studies. This review re-
vealed insights on methods, revealing that genomic,
ECG, and medical record data have been targeted due
to the unique patterns and use of auxiliary informa-
tion. This led us to outline the challenges and pro-
pose actionable suggestions to enhance practical as-
sessments in healthcare.
Our contribution introduces a perspective on a de-
anonymization attack that leverages insights by out-
lining potential strategies and contextualizing them
to data types. We explore the investigation of
telemedicine, raising new privacy challenges caused
by the digital shift in healthcare and integration of
real-time data from various sources, resulting in the
need for more robust data protection measures based
on privacy-preserving mechanisms such as DP and
ICISSP 2025 - 11th International Conference on Information Systems Security and Privacy
604

tuning models to customizable privacy budgets while
keeping data utility, utilization of techniques like
GANs (Yoon et al., 2020) and Variational Autoen-
coders (VAEs) (Yang et al., 2023) to enhance syn-
thetic data generation as well as homomorphic en-
cryption delivering better protection for health data
(Regazzoni et al., 2024). Our findings outline a
roadmap for investigations into the privacy and con-
fidentiality of health information. We emphasize the
ethical and secure sharing of health data aligned with
anonymization techniques. We highlight a fine line
between data utility and protection in today’s digital
age, and our findings set a foundation for ongoing re-
search to maintain this balance.
This review introduced strategies for future works;
as a part, we emphasize expanding our investiga-
tion on developing a detailed benchmarking frame-
work to evaluate de-anonymization techniques com-
prehensively. This includes a comparative evalua-
tion of methodologies, experimental validation, and
reproducibility assessments to bridge gaps in cur-
rent research, go beyond re-identification, and use
de-anonymization for fraudulent activity detection
within healthcare systems.
Our extended version will further elaborate on
these aspects, providing a more in-depth analysis of
de-anonymization challenges, research gaps, and in-
novative solutions to advance the field.
REFERENCES
Al-Azizy, D., Millard, D., Symeonidis, I., O’Hara, K., and
Shadbolt, N. (2016). A literature survey and classifica-
tions on data deanonymisation. In Risks and Security
of Internet and Systems. Springer International.
Antoniou, A., Dossena, G., MacMillan, J., Hamblin, S.,
Clifton, D., and Petrone, P. (2022). Assessing the
risk of re-identification arising from an attack on
anonymised data.
Ayday, E. and Humbert, M. (2017). Inference attacks
against kin genomic privacy. IEEE Security & Pri-
vacy, 15(5):29–37.
Ayoz, K., Ayday, E., and Cicek, A. E. (2021). Genome Re-
construction Attacks Against Genomic Data-Sharing
Beacons. PoPETs, 2021(3):28–48.
Bayardo, R. J. and Agrawal, R. (2005). Data privacy
through optimal k-anonymization. In 21st Interna-
tional conference on data engineering (ICDE’05),
pages 217–228. IEEE.
BEUC (2023). Consumer Attitudes to Health Data Sharing:
Survey Results from Eight EU Countries. Technical
Report BEUC-X-2023-051, BEUC, European Con-
sumer Organisation.
Bhattacharya, M., Roy, S., Chattopadhyay, S., Das, A. K.,
and Shetty, S. (2023). A comprehensive survey on
online social networks security and privacy issues:
Threats, machine learning-based solutions, and open
challenges. Security and Privacy, 6(1):e275.
Branson, J., Good, N., Chen, J.-W., Monge, W.,
Probst, C., and Emam, K. E. (2020). Evaluating
the re-identification risk of a clinical study report
anonymized under ema policy 0070 and health canada
regulations. Trials.
Dimitrievski, A., Loncar-Turukalo, T., and Trajkovik, V.
(2023). Securing patient information in connected
healthcare systems in the age of pervasive data col-
lection. In 2023 IEEE MeditCom.
Ding, X., Zhang, L., Wan, Z., and Gu, M. (2010). A brief
survey on de-anonymization attacks in online social
networks. In 2010 CASoN. IEEE.
Edge, M. D. and Coop, G. (2020). Attacks on genetic pri-
vacy via uploads to genealogical databases. eLife,
9:e51810.
Emam, K. E., Jonker, E., Arbuckle, L., and Malin, B.
(2011). A systematic review of re-identification at-
tacks on health data. PLoS ONE, 6(12):e28071.
European Union (2016). Regulation (eu) 2016/679 general
data protection regulation. Official Journal of the Eu-
ropean Union. eur-lex.europa.eu/eli/reg/2016/679/oj.
Farzanehfar, A., Houssiau, F., and de Montjoye, Y.-A.
(2021). The risk of re-identification remains high
even in country-scale location datasets. Patterns,
2(3):100204.
Ghazarian, A., Zheng, J., Struppa, D., and Rakovski, C.
(2022). Assessing the reidentification risks posed by
deep learning algorithms applied to ecg data. IEEE
Access, 10:68711–68723.
Gymrek, M., McGuire, A. L., Golan, D., Halperin, E., and
Erlich, Y. (2013). Identifying Personal Genomes by
Surname Inference. Science, 339(6117):321–324.
Henriksen-Bulmer, J. and Jeary, S. (2016). Re-identification
attacks—a systematic literature review. International
Journal of Information Management, 36:1184–1192.
Hong, S., Wang, C., and Fu, Z. (2020). Cardioid: Learning
to identification from electrocardiogram data. Neuro-
computing, 412:11–18.
Humbert, M., Huguenin, K., Hugonot, J., Ayday, E.,
and Hubaux, J.-P. (2015). De-anonymizing Ge-
nomic Databases Using Phenotypic Traits. PoPETs,
2015(2):99–114.
IBM Security (2023). Cost of a data breach report 2023.
Technical report, IBM Security.
Ji, S., Gu, Q., Weng, H., Liu, Q., Zhou, P., Chen, J., Li, Z.,
Beyah, R., and Wang, T. (2020). De-Health: All Your
Online Health Information Are Belong to Us. In 2020
IEEE 36th International Conference on Data Engi-
neering (ICDE), pages 1609–1620, Dallas, TX, USA.
IEEE.
Ji, S., Wang, T., Chen, J., Li, W., Mittal, P., and Beyah,
R. (2019). De-sag: On the de-anonymization of
structure-attribute graph data. IEEE Transactions on
Dependable and Secure Computing, 16(4):594–607.
Kitchenham, B. (2004). Procedures for performing sys-
tematic reviews. Keele, UK, Keele University,
33(2004):1–26.
De-Anonymization of Health Data: A Survey of Practical Attacks, Vulnerabilities and Challenges
605

Lange, L., Schreieder, T., Christen, V., and Rahm,
E. (2023). Privacy at risk: Exploiting similari-
ties in health data for identity inference. CoRR,
abs/2308.08310.
Lee, W.-H., Liu, C., Ji, S., Mittal, P., and Lee, R. B.
(2017). Blind de-anonymization attacks using social
networks. In Proceedings of the 2017 on Workshop on
Privacy in the Electronic Society, pages 1–4.
Li, N., Li, T., and Venkatasubramanian, S. (2006).
t-closeness: Privacy beyond k-anonymity and l-
diversity. In 2007 IEEE 23rd international conference
on data engineering, pages 106–115. IEEE.
Lippert, C., Sabatini, R., Maher, M. C., Kang, E. Y., Lee,
S., et al. (2017). Identification of individuals by trait
prediction using whole-genome sequencing data. Pro-
ceedings of the National Academy of Sciences.
Liu, Y., Wan, Z., Xia, W., Kantarcioglu, M., Vorobeychik,
Y., Clayton, E. W., Kho, A., Carrell, D., and Malin,
B. A. (2018). Detecting the presence of an individual
in phenotypic summary data. In AMIA Annual Sym-
posium Proceedings, pages 760–769.
Lu, G., Li, K., Wang, X., Liu, Z., Cai, Z., and Li, W. (2024).
Neural-based inexact graph de-anonymization. High-
Confidence Computing, 4(1).
Machanavajjhala, A., Kifer, D., Gehrke, J., and Venkita-
subramaniam, M. (2007). l-diversity: Privacy beyond
k-anonymity. Acm TKDD, 1(1):3–es.
Malin, B. and Sweeney, L. (2004). How (not) to pro-
tect genomic data privacy in a distributed network:
Using trail re-identification to evaluate and design
anonymity protection systems. Journal of Biomedical
Informatics, 37(3):179–192.
Min-Gu Kim, Hoon Ko, S. B. P. (2020). A study on user
recognition using 2d ecg based on ensemble of deep
convolutional neural networks. Journal of Ambient In-
telligence and Humanized Computing.
Mitchell, A. R. J., Ahlert, D., Brown, C., Birge, M., and
Gibbs, A. (2023). Electrocardiogram-based biomet-
rics for user identification – using your heartbeat as a
digital key. Journal of Electrocardiology, 80:1–6.
Narayanan, A. and Shmatikov, V. (2008). Robust de-
anonymization of large sparse datasets. In 2008 IEEE
Symposium on Security and Privacy. IEEE.
Nasr, M., Shokri, R., and Houmansadr, A. (2019). Compre-
hensive privacy analysis of deep learning: Passive and
active white-box inference attacks against centralized
and federated learning. In 2019 IEEE SP.
Prada, S. I., Gonz
´
alez-Mart
´
ınez, C., Borton, J., et al.
(2011). Avoiding disclosure of individually identifi-
able health information: a literature review. SAGE
Open, 1(3):2158244011431279.
Ravindra, V. and Grama, A. (2021). De-anonymization At-
tacks on Neuroimaging Datasets. In Proceedings of
the 2021 International Conference on Management of
Data, pages 2394–2398, Virtual Event China. ACM.
Regazzoni, F., Acs, G., Palmieri, P., et al. (2024). Secured
for health: Scaling up privacy to enable the integration
of the european health data space. In 2024 Design, Au-
tomation & Test in Europe Conference & Exhibition
(DATE), pages 1–4. IEEE.
Saxena, R., Arora, D., Nagar, V., and Chaurasia, B. K.
(2024). Blockchain transaction deanonymization us-
ing ensemble learning. Multimedia Tools and Appli-
cations, pages 1–30.
Schmidt, P., Reiss, A., Duerichen, R., Marberger, C., and
Van Laerhoven, K. (2018). Introducing wesad, a mul-
timodal dataset for wearable stress and affect detec-
tion. In Proceedings of the 20th ACM ICMI, pages
400–408.
Shokri, R., Stronati, M., Song, C., and Shmatikov, V.
(2017). Membership inference attacks against ma-
chine learning models. In 2017 IEEE Symposium on
Security and Privacy (SP), pages 3–18. IEEE.
Shringar-pure, S. S. and Bustamante, C. D. (2015). Pri-
vacy Risks from Genomic Data-Sharing Beacons. The
American Journal of Human Genetics, 97(5):631.
Sweeney, L. (1997). Weaving technology and policy to-
gether to maintain confidentiality. The Journal of Law,
Medicine & Ethics, 25(2-3):98–110, 82.
Sweeney, L. (2002). k-anonymity: A model for protecting
privacy. International Journal of Uncertainty, Fuzzi-
ness, and Knowledge-Based Systems.
Thenen, N. V., Ayday, E., and Cicek, A. E. (2019).
Re-identification of individuals in genomic data-
sharing beacons via allele inference. Bioinformatics,
35(3):365–371.
U.S. Congress (1996). Health Insurance Portability and Ac-
countability Act of 1996 (HIPAA). Public Law 104-
191, 110 Stat. 1936.
Venkat-esaramani, R., Malin, B. A., and Vorobeychik, Y.
(2021). Re-identification of individuals in genomic
datasets using public face images. Science Advances,
7(47):eabg3296.
Wan, Z., Vorobeychik, Y., Xia, W., Liu, Y., et al. (2021).
Using game theory to thwart multistage privacy in-
trusions when sharing data. Science Advances,
7(50):eabe9986.
Wu, M., Zhang, X., Ding, J., Nguyen, H., Yu, R., Pan, M.,
and Wong, S. T. (2020). Evaluation of inference at-
tack models for deep learning on medical data. arXiv
preprint arXiv:2011.00177.
Yang, R., Ma, J., Miao, Y., and Ma, X. (2023). Privacy-
preserving generative framework for images against
membership inference attacks. IET Communications,
17(1):45–62.
Yin, H., Liu, Y., Li, Y., Guo, Z., and Wang, Y. (2023).
Defeating deep learning based de-anonymization at-
tacks with adversarial example. Journal of Network
and Computer Applications, 220.
Yoon, J., Drumright, L. N., and Van Der Schaar, M. (2020).
Anonymization through data synthesis using genera-
tive adversarial networks. IEEE J-BHI, 24(8).
Zhang, Z., Yan, C., and Malin, B. A. (2022). Membership
inference attacks against synthetic health data. Jour-
nal of Biomedical Informatics, 125:103977.
ICISSP 2025 - 11th International Conference on Information Systems Security and Privacy
606
