
A Multimodal Approach to Research Paper Summarization
Pranav Bookanakere
1
, Syeda Saniya
1
, Syed Munzer Nouman
1
, Pramath S
1
and Jayashree Rangareddy
2
1
Department of Computer Science and Engineering, PES University, Bangalore, India
2
Department of Computer Science and Engineering - AI/ML, PES University, Bangalore, India
Keywords:
Research Paper, Machine Learning, T5, Transformers, Self-Attention Mechanism, Feed Forward Neural
Networks (FNNs), Cross-Attention Mechanism, Vision Transformer (ViT).
Abstract:
As the amount of academic research in the medical field has been growing exponentially, being able to un-
derstand and extract important information from these research papers has become all the more challenging.
Researchers, students, and professionals often find it hard to navigate through medical-based research papers
that contain complex images and textual information. Most summarization tools that already exist have limited
effectiveness and cannot handle the multimodal nature of complex research papers. This paper addresses the
need for an all-round approach to effectively generate summaries, taking key information from both the text as
well as the complex images present in research papers. Our approach can generate section-wise summaries of
the text and also generate context-based image descriptions with high levels of accuracy. By putting together
advanced Natural Language Processing (NLP) and multimodal (T5, Llava) techniques, this system is able
to generate comprehensive and concise summaries of complex research papers. This work demonstrates the
potential of multimodal AI models to improve research comprehension and provide deeper understanding of
complex subjects in the medical field.
1 INTRODUCTION
The exponential growth of academic research in the
medical field is creating an abundance of information,
which is making it very hard for researchers, profes-
sionals, and students to review the contents of the re-
search papers efficiently. These papers often consist
of multiple pages and include complex terminology
and graphical data, which takes a lot of time and en-
ergy to comprehend. This is further complicated by
trying to make sense of the textual content as well as
the visual content that comes along with it, as both
aspects are vital for the better context. Reading be-
came more important as well as more difficult as the
complexity of medical research papers increased. In-
terdisciplinary research became the norm, and it has
markedly grown in importance with every research,
growing the need for a stronger and deeper under-
standing of research papers.
According to a study by two neuroscientists at
the Karolinska Institute in Stockholm, who scoured
through 700,000 English-language abstracts pub-
lished between 1881 and 2015 in 122 leading biomed-
ical journals, they posted that jargon-heavy phrasing
is not the only problem with modern-day scientific re-
search papers, but there has also been an increase in
“general scientific jargon” which refers to multisylla-
ble words that have non-technical meanings but have
become part of the standard lexicon of modern-day
science papers (Thompson, 2017). Researchers be-
longing to other fields also end up suffering because
a lot of the knowledge ends up getting trapped within
the fields as the language and images in medical sci-
entific papers prove to be very difficult to understand
and interpret.
Artificial Intelligence (AI) and Natural Language
Processing (NLP) have shown significant progress in
trying to automate the process of summarizing text
accurately through various advanced models and sys-
tems. However, there is a crucial element missing in
most existing models and systems, and that is the abil-
ity to capture information through the visual content
present in these research papers. Images present in
research papers provide better context and a deeper
understanding of the concept that is being explained
through the textual content. We propose a novel mul-
timodal system that is capable of extracting and in-
terpreting both text and images in medical-based re-
950
Bookanakere, P., Saniya, S., Nouman, S. M., Pramath, S. and Rangareddy, J.
A Multimodal Approach to Research Paper Summarization.
DOI: 10.5220/0013277700003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 2: VISAPP, pages
950-957
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

search papers to bridge this gap.
Our system aims to harness the combined power
of NLP techniques and multimodal learning to im-
prove the comprehension and summarization power
of research papers in the medical field. We utilize a
comprehensive dataset that includes behavioral pat-
terns, self-reported symptoms and demographic data
to create a detailed profile of an individual’s mental
state. By applying techniques such as Transformers,
Feed-Forward Neural Networks, we strive to enhance
the accuracy and comprehensiveness of medical re-
search paper summaries.
The integration of T5 as the text summarization
model LLaVA-1.5 7B as the image description gen-
eration model ensures that an extremely comprehen-
sive, concise, and accurate summary of the research
paper is developed. The summary takes into account
the textual content, the image description, as well as
the context that the image links to in order to gener-
ate an overall summary that is not only accurate but
also provides a deep understanding of the subject the
research paper is trying to address. A holistic view
of the research paper is offered through our system,
which significantly helps reduce time and effort in try-
ing to comprehend complex medical research papers.
This system exhibits the effectiveness and power
of multimodal AI in extraction as well as concise
summarization of crucial information in complex
medical research papers.
2 RELATED WORK
With the exponential growth in the number of re-
search papers in every domain of study, researchers
often find it extremely hard to keep up with new de-
velopments and sift through the wide variety of arti-
cles to gain an deeper understanding of crucial con-
cepts. This brings out the importance for a more au-
tomated approach to generate concise, accurate and
comprehensive method to generate summaries. Ex-
tractive summarization ensures high accuracy by se-
lecting important sentences but may develop incoher-
ent summaries. Abstractive summarization (Gupta
and Gupta, 2018) (Lin and Ng, 2019), on the other
hand, generates concise and readable summaries but
may lose key facts and important information (Shukre
et al., 2023).
BART (Bidirectional and Auto-Regressive Trans-
former) a denoising autoencoder for pretrain-
ing sequence-to-sequence models, uses a standard
Transformer-based neural machine translation archi-
tecture which despite it’s simplicity, helps to gen-
eralize BERT and other recent pre-training schemes
(Mike Lewis and et al., 2020). The T5 model
is a flexible and potent transformer-based language
model that uses a uniform framework to work on
multiple natural language processing tasks. The T5
model effectively uses the transformer architecture
to encode, decode and generate outputs using self-
attention mechanisms, feed-forward networks, and
masked decoding (Shukre et al., 2023).
Abstractive Text Summarization (Gupta and
Gupta, 2018) (Lin and Ng, 2019) is an important sum-
marization task which rephrases the input text into a
short version summary while trying to preserve the
important semantics (Guan et al., 2021). Automatic
Text Summarization (ATS) is a rapidly growing field
that aims to reduce the time and effort that is put in
by readers, by automatically generating summaries
of large volumes of text using hybrid extractive (Liu,
2019) (Zhong et al., 2020) and abstractive techniques
for summarization (Khan et al., 2023). The most
commonly used architecture for applying sequence-
to-sequence models is the encoder-decoder architec-
ture. Regardless of the advancements in ATS, long-
term dependency handling is still not as efficient as is
needed (Alomari et al., 2021).
Another hybrid approach that used the transformer
model in combination with the Luhn algorithm to
summarize text extracted by Tesseract OCR proved to
provide a good level of accuracy and comprehension
abilities. A comparison was drawn between this hy-
brid model and the already existing abstractive (Gupta
and Gupta, 2018) (Lin and Ng, 2019) model using
ROGUE metrics; the fine-tune model got the high-
est ROUGE score during evaluation; the ROUGE-1,
ROUGE-2 and ROUGE-L score was 57%, 43% and
42% respectively (Zachary et al., 2022).
The SCICAP dataset was a figure caption dataset
based on computer science research papers that was
used to build an end-to-end neural network-based
model framework for automatically generating infor-
mative and high-quality captions for scientific images
(Hsu et al., 2021). Another unique approach involved
the use of cross-modal learning in order to gener-
ate more precise captions for scientific images and
showed significant results. This cross-modal learn-
ing technique used sequence-level learning model for
accurate figure captioning along with another unique
approach of treating figure captioning as a text sum-
marization task to leverage automated summarization
models like PEGASUS (Chen et al., 2019) (Huang
et al., 2023).
An encoder (Convolutional Neural Network
(CNN)) along with a decoder (Recurrent Neural Net-
work (RNN)) based framework would be able to ex-
tract visual features using the encoder and develop de-
A Multimodal Approach to Research Paper Summarization
951

scriptive text for the same. This CNN-RNN based
framework, along with a joint image/text context
cascade mechanism that treats the process of medi-
cal imaging annotation as a multi-label classification
task. But the issue with reports generated using this
method was incoherence and difficulty to comprehend
(Shin et al., 2016). A development along these lines
was proposed by adding an enhancement using an
Auxillary Attention Sharpening (AAS) module to be
able to automatically generate the medical image cap-
tions. This method was posing the issue of a word
limit of only 59 words along with a topic limitation of
5 topics only (Zhang et al., 2017).
In a recent development in this field of medical
image captioning, a hierarchical co-attention based
model to generate multi-task medical imaging reports
was proposed which contained a vast variety of het-
erogeneous data including short labels, as Medical
Text Indexers (MTIs) (James G. Mork, 2013) tags,
long paragraph of text as findings, and a summary of
findings as impression. This model is able to attend to
the image while also being able to predict the tags us-
ing visual and semantic information. It can also gen-
erate long sequence sentences using the LSTM frame-
work. The model proved to be prone to false posi-
tives due to interference of irrelevant tags (Jing et al.,
2018). A development on this same methodology was
proposed by building a hierarchical reinforced medi-
cal image report generation model, introducing rein-
forcement learning and template based language gen-
eration. But it’s reliance on RNN as the decoder archi-
tecture prevented parallel computation and difficulty
in generalizing on other applications (Li et al., 2018).
Another model used a hierarchical neural network
model architecture which used a reinforced trans-
former methodology to try to overcome some of
the aforementioned issues. Some improvements in-
volved, using a transformer model to be able to cap-
ture long-term dependencies in images as well as sen-
tences, implementing a bottom-up attention mecha-
nism using the pretrained DenseNet model, and the
additional use of reinforcement learning based train-
ing methods for preventing exposure bias. But since
the the model used the DenseNet model pretrained
on an exclusive images of chest X-rays limiting it’s
ability to generalize to other medical images (Yux-
uan Xiong, 2019).
3 DATASET
In this work, We have fine-tuned both the T5 (Raf-
fel et al., 2019) and Llava (Li et al., 2024) models,
to specialize in the task of summarizing/captioning
medical research content. For the T5 model we used
the PubMed Article Summarization Dataset since its
large collection of biomedical articles and coherent
summaries would be convenient to develop a deeper
understanding of the medical content in the research
paper. Gradient clipping with learning rate scheduling
optimize it for better performance. The Llava model
was fine-tuned on the MedPix 2.0 dataset which is the
only dataset specially curated and built for medical
image captioning purposes. With medical images ac-
companied by descriptive captions and further details
in research articles, the model could capture images
and figures with their relative contexts and describe
them accurately. The outcome is a whole multimodal
model that synthesizes image and text seamlessly to
produce precise, coherent and concise summaries.
4 IMPLEMENTATION
The method we used in building this system is by cre-
ating a pipeline flow for both text, table as well as im-
age summarization tasks as shown in “Fig. 1”. Sepa-
rate summaries for text and corresponding images are
generated by the system, which are then combined to
generate a more concise, comprehensive and under-
standable final summary of the research paper. The
text summarization pipeline extracts, processes and
then cleans the text data to forward it for summariza-
tion by the T5 model.
4.1 Text Summarization
The text summarization pipeline extracts, processes
and then cleans the text data in the PDF format. This
final prepared text if then forwarded for summary
generation by the fine-tuned T5 model.
4.1.1 PDF Preprocessing and Text Extraction
In this study, we have utilized a combination
of libraries, including pdfplumber, PyPDF2, and
PyMuPDF, for the purpose of extracting the text from
the PDFs. With the incorporation of this step it is en-
sured that the system is capable of handling both the
simple text as well as more complex representations,
such as multi-column text and embedded tables. The
division of pages into two equal halves in standard re-
search papers is identified, and the text in each section
is carefully extracted and stored. The extract table
function from pdfplumber handles the extraction and
conversion to text-based format of the tabular data.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
952
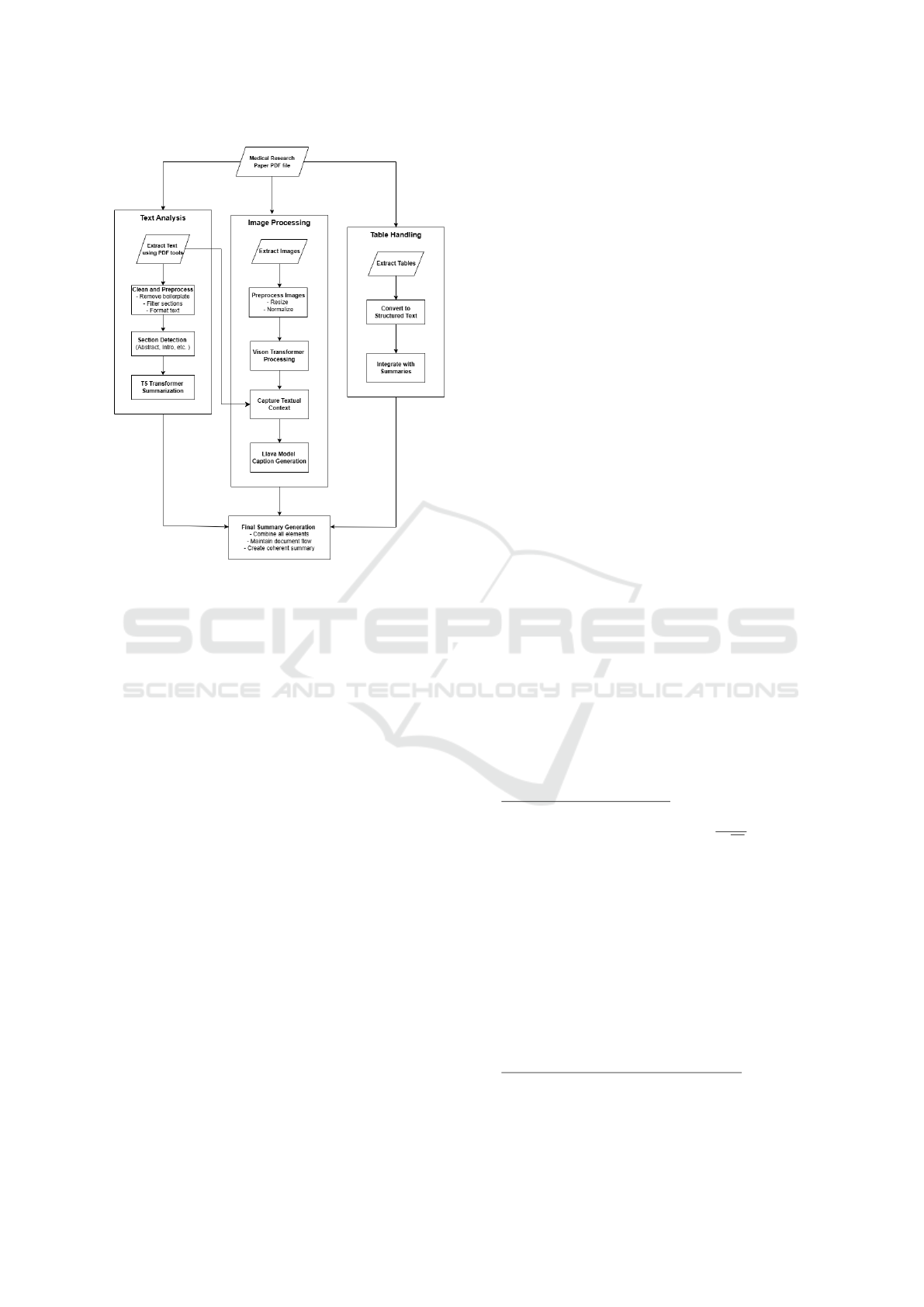
Figure 1: Model Pipeline Flow.
4.1.2 Preprocessing and Text Cleaning
The extracted in the previous steps undergoes multi-
ple preprocessing tasks to prepare it for summariza-
tion:
• Text Filtering: The unnecessary parts, such as
the text present in the paper after the References
section and boilerplate elements, are discarded.
To effectively understand why the text present in
the paper after the References section (e.g. Ap-
pendices) is deemed unnecessary for summariza-
tion, we considered a few important points.
Some possible boilerplate elements are Appen-
dices, Acknowledgements, Author Biographies,
Funding and Conflict of Interest Statements and
Supplementary References or Notes. The primary
goal of a research paper summarizer is to con-
dense the core arguments, findings and conclu-
sions of the paper. Appendices and other sections
after References typically contain supplementary
or non-essential information that do not directly
contribute to the paper’s main narrative. Author
acknowledgements or funding details are impor-
tant for clarity and providing credit to the nec-
essary sources, but are irrelevant to the summa-
rizer’s task of extracting the essence of the re-
search. Many of these elements are extensions
of data or methods already covered in the main
body and must be omitted to avoid redundancy.
Removing text after References standardizes the
input across diverse papers, making the summa-
rization process more consistent, considering that
different papers follow different structures to be-
gin with.
• Section Identification: Further segmentation of
the extracted text was done by utilizing head-
ing patterns based on Roman Numerals(e.g., “I.
Introduction”) determining the section bound-
aries. Using this regular expression meaningful
sections/segments were generated, such as “Ab-
stract”, “Introduction”, etc.
• Text Reduction: The key section (“Abstract”)
was identified to truncate the content and all other
redundant data before and after the main body of
the paper was removed.
4.1.3 Summarization Process Using
Transformer Models
In order to generate the summaries for these sections
we employed the sequence-to-sequence Transformer
model, T5 (Text-to-Text Transfer Transformer). This
model is very well suited for text processing tasks
such as text generation, summarization, etc. The
model leverages an encoder-decoder architecture to
generate concise and comprehensive summaries. The
encoders are responsible for generating dense repre-
sentations of the raw text fed to them. The multi-
head self-attention layer in the encoders helps weigh
the different parts of the input to determine relevance
and dependency. This helps maintain the important
parts of the text. The Feed-forward neural network
(FNN) works on building more abstract representa-
tions to gain deeper understanding.
Self-Attention Mechanism:
Attention(Q, K, V ) = softmax
QK
T
√
d
k
V (1)
Where,
• Q, K, and V are the query, key, and value matrices.
• d
k
is the dimension of the key vector.
• QK
T
is the dot product of the query and key ma-
trices.
• softmax is the softmax function applied to the
scaled dot product.
• V is the value matrix that is multiplied with the
attention weights.
Feed-Forward Neural Network (FNN):
FNN(x) = ReLU(xW
1
+ b
1
)W
2
+ b
2
(2)
Where,
A Multimodal Approach to Research Paper Summarization
953

• x is the input to the FNN.
• W
1
, W
2
are weight matrices for the two linear
transformations.
• b
1
, b
2
are bias vectors.
• ReLU is the rectified linear unit activation func-
tion applied element-wise.
• The final output is a linear transformation of the
intermediate hidden layer.
The decoders use hidden states to predict the next
word by using the already generated output and the
encoder representations. They help generate a final,
concise summary.
Cross-Attention in Decoder:
CrossAttention(Q
d
, K
e
, V
e
) = softmax
Q
d
K
T
e
√
d
k
V
e
(3)
Where,
• Q
d
is the query matrix from the decoder input.
• K
e
, V
e
are the key and value matrices from the en-
coder output.
• d
k
is the dimension of the key vector.
• Q
d
K
T
e
is the dot product of the decoder queries
and encoder keys.
• softmax is the softmax function applied to scale
the dot product and generate attention weights.
• V
e
is the encoder output value matrix, weighted by
the attention weights.
Each section (e.g., Abstract, Introduction) is sum-
marized independently by the T5 model developing
concise summaries. The summaries retain all the crit-
ical and important information from the original pa-
pers, yet help in significantly reducing the length.
Word Prediction Probability:
P(word) = softmax(xW + b) (4)
Where,
• P(word) represents the predicted probability dis-
tribution over the vocabulary for the next word.
• x is the input vector from the decoder (usually
the output of the final decoder layer or a previous
word embedding).
• W is the weight matrix that transforms the input
vector into the output space of vocabulary size.
• b is the bias term.
• softmax is the softmax function, which converts
the output logits into a probability distribution.
4.1.4 Combining Tables with Text Summaries
Tables offer a lot of important information and help
gain a deeper understanding of the research paper.
Thus, their integration into the summaries is integral.
This is done by using pdfplumber model to extract
the tables from the original research paper. The ex-
tracted tables are then converted into a structured for-
mat i.e., plain text and gets saved separately.
Each a particular section is being summarized, the
systems refers back to the structured representations
of the corresponding tables to draw inferences and
make the summary more meaningful and insightful.
4.2 Image Description Generation
Detailed descriptions of the images in the research pa-
per are generated using the Llava 7b model which is
trained for complex image-to-text tasks, Images are
first extracted from the PDF and preprocessed before
being fed to the model for description generation.
4.2.1 Image Preprocessing
During the PDF preprocessing stage, the figures that
are extracted from the research paper PDF are cleaned
and prepared for feeding to the Llava model:
• Figure Extraction: Using the pdfplumber
model, figures and charts are extracted from the
original research paper. They are stored as in-
dependent image files associated with their cor-
responding figure numbers in the text.
• Image Preparation for Model: The extracted
images are further preprocessed by resizing and
normalizing them based on the input requirements
of the Llava model. The image is further con-
verted into a tensor format, which would serve as
the input to the model for description generation.
4.2.2 Transformer-Based Visual Processing
The input image is split into patches which are then
flattened to form 1D vectors. The projection of these
vectors in a lower dimensional space generates patch
embeddings.
z
patch
= Linear(Flatten(x
patch
)) + PositionalEncoding
(5)
Where,
• x
patch
is the image patch.
• z
patch
is the embedding of the patch after linear
transformation.
• PositionalEncoding is added to the embeddings to
retain spatial information.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
954
The Vision Transformer (ViT) takes a sequence
of these patch embeddings as input and passes
them through layers of the Transformer to extract
meaningful visual features. The multi-head self-
attention mechanism and Feed-forward Neural Net-
works (FNNs) help the model in learning high-level
representations of the images.
4.2.3 Multimodal Interaction
The embeddings generated by the Vision Transformer
(ViT) are integrated with the textual inputs which in-
clude the text surrounding the images as well as di-
rect references to the images in the text like “Figure
1 shows. . . ” or “Refer to Fig. 3.” with the help of
the cross-attention mechanism. This helps in adding
much greater context to the caption for the image, en-
suring the caption is both more accurate and compre-
hensive.
4.2.4 Image-to-Text Captioning
After the integration of both the images and their re-
spective textual content, the language decoders gen-
erate detailed and descriptive caption using the cross-
attention mechanism and token prediction, similar to
the text summary generation.
4.3 Final Summary Generation
After the generation of text summaries of all the rel-
evant sections and image descriptions through textual
references and visual understanding and incorpora-
tion of tabular data, the system is capable of concate-
nating them all together in order to develop a concise
and comprehensive summary. The summaries of each
of the sections are combined in the same order as they
appear in the original research paper. The natural flow
of the text, figures and tables is maintained while gen-
erating the final paper summary.
5 RESULTS AND DISCUSSIONS
The performance results of each of the individually
fine tuned models was observed and our combined
pipeline model’s performance was compared using
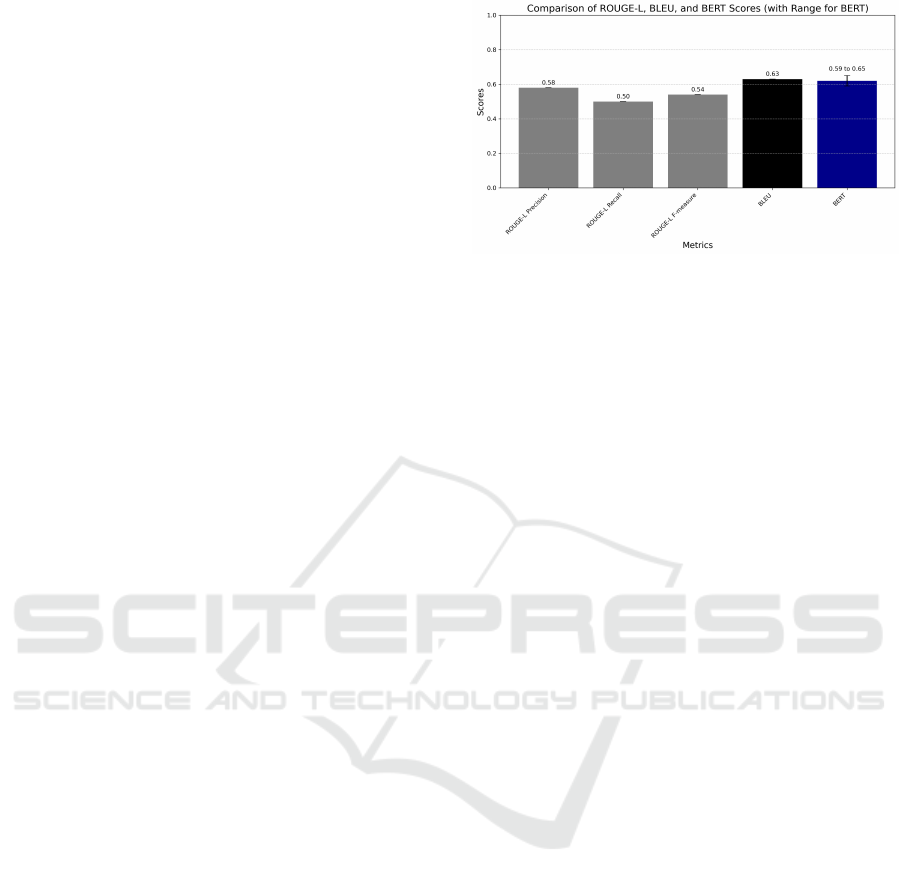
three different metrics as shown in “Fig. 2”, which
has been discussed below:
5.0.1 ROUGE Score Analysis
The overall multimodal model achieved an average
ROGUE score of 0.54. This score surpasses the es-
tablished method of summarization using an OCR-
Figure 2: Comparison of Metrics.
based hybrid summarizer as indicated by [14] as well
as other major existing summarization approaches
which are present in our References. Thus, proving
the ability of the overall model to capture the logical
overlap with other relevant medical research articles
and summarize them effectively.
5.0.2 BLEU Score Evaluation
Using the BLEU score evaluation method, we
achieved an average BLEU score of 0.64, which, in
comparison to all existing medical-based research pa-
per summarizers, proves to be a significant improve-
ment. This score suggests that the summarizer is ca-
pable of capturing relevant details as well as maintain-
ing the sentence structure and semantics. The model
is able to maintain the language and flow of the re-
search article while effectively summarizing it.
5.0.3 Limitations of BLEU and ROUGE
• BLEU and ROGUE score analysis is an effective
method to determine the overlap with the origi-
nal paper determining the similarity between the
original research article and the generated sum-
mary. But the issue with this approach of deter-
mining the performance of the model is that it
does not effectively determine it’s ability to cap-
ture the meaning conveyed through the article, es-
pecially in more factually heavy articles.
• Thus we chose to opt for the BERTScore method
for a more specific and nuanced method to de-
termine the ability of the model to retain fac-
tual correctness and capture the true meaning con-
veyed. The BERTScore method leverages contex-
tual embeddings to assess the semantic alignment
between the original paper and the generated sum-
mary.
A Multimodal Approach to Research Paper Summarization
955

5.0.4 BERTScore Evaluation
Using the BERTScore method we achieved a
BERTScore range of 0.59 to 0.65, which in the con-
text of medical research paper summarization proves
to be impressive. This score proves that the model is
able to retain the semantic structure and language of
the author while also being able to retain the factual
information that is relevant. Most medical literature
is factual-heavy and includes complex terminology,
thus, this score proves the ability of our model to cap-
ture the essential information and meaning conveyed
by the medical research article.
This performance is calculated as an average over
10,000 validation rows in our dataset. Notably, we
observed BERTScore values exceeding 0.8 for many
individual rows, highlighting the model’s exceptional
summarization capability in many cases.
5.0.5 Discussion on Performance in Medical
Context
The results as seen in Table I and the discussions
above are indicative of the fact that our model is ca-
pable of handling the domain language and terminol-
ogy specific to each medical research paper, which
is extremely crucial when generating summaries that
are descriptive yet comprehensive. The combina-
tion of lexical-based and semantic similarity metrics,
highlights the ability the quality of the summarizer
model, by determining the surface-level overlap and
the deeper meaning alignment.
Table 1: Comparison of ROUGE-L Scores.
Model ROUGE-L Score
Base Paper [14] 0.42
Our Summarization Model 0.54
6 CONCLUSION AND FUTURE
WORK
This paper introduces an innovative multimodal ap-
proach for summarizing medical research papers,
moving beyond traditional text-only summaries to
create comprehensive and contextually rich outputs
that integrate both textual and visual data. By lever-
aging the combination of fine-tuned T5 and LLaVA
models, this approach enhances medical research pa-
per summarization and captioning. The T5 model,
fine-tuned on the PubMed Article Summarization
Dataset, effectively condenses complex biomedical
texts into concise yet informative summaries. Mean-
while, the LLaVA model, trained on MedPix 2.0, gen-
erates captions that capture visual information from
medical images, ensuring alignment with their textual
context. This multimodal framework bridges the gap
between textual, tabular and visual data, significantly
improving the comprehension and summarization of
complex biomedical literature for a diverse range of
users.
This approach represents a transformational shift
in medical documentation processing by integrating
text and images into a unified summarization system.
Beyond benefiting researchers, this framework also
lays the foundation for automated tools that can assist
doctors, academicians and policymakers in efficiently
keeping up with the latest advancements while mini-
mizing time and effort.
Looking ahead, future work will focus on devel-
oping a unified architecture capable of processing text
and images simultaneously, eliminating the need for
separate models and ensuring better coherence across
modalities. Additionally, long-context models will be
explored to handle extensive biomedical documents,
ensuring detailed yet concise summaries. The project
can also be extended by incorporating diverse datasets
to improve generalization and refining the model for
better adaptability across various biomedical research
domains and integrating other forms of data like text-
based charts and tables. Further enhancements could
include advanced attention mechanisms to strengthen
text-image integration fidelity within summaries. An-
other key direction is adaptive summarization, allow-
ing the system to generate summaries tailored to dif-
ferent user expertise levels, ensuring accessibility for
both specialists and general readers.
7 ABBREVIATIONS
In this section, we have tried to list and explain some
abbreviations that have not been explained in the pa-
per above:
1. NLP - Natural language processing (NLP) is
a subfield of computer science and artificial intelli-
gence (AI) that uses machine learning to enable com-
puters to understand and communicate with human
language.
2. BERT - BERT language model is an open
source machine learning framework for natural lan-
guage processing (NLP). It stands for Bidirectional
Encoder Representations from Transformers, is based
on transformers, a deep learning model in which ev-
ery output element is connected to every input ele-
ment, and the weightings between them are dynami-
cally calculated based upon their connection.
3. OCR - Optical Character Recognition (OCR)
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
956

is the process that converts an image of text into a
machine-readable text format.
4. CNN - A Convolutional Neural Network
(CNN) is a type of deep learning algorithm that is par-
ticularly well-suited for image recognition and pro-
cessing tasks. It is made up of multiple layers, in-
cluding convolutional layers, pooling layers, and fully
connected layers.
5. RNN - A recurrent neural network or RNN is
a deep neural network trained on sequential or time
series data to create a machine learning (ML) model
that can make sequential predictions or conclusions
based on sequential inputs.
6. FNN - A feedforward neural network is a type
of neural network where information flows in one di-
rection from the input to the output layers, without
cycles or loops.
7. LSTM - LSTM (Long Short-Term Memory) is
a recurrent neural network (RNN) architecture widely
used in Deep Learning. It excels at capturing long-
term dependencies.
8. ROUGE - ROUGE (Recall-Oriented Under-
study for Gisting Evaluation), is a set of metrics and a
software package specifically designed for evaluating
automatic summarization, but that can be also used
for machine translation.
9. BLEU - The acronym BLEU refers to a “Bilin-
gual Evaluation Understudy”, and it’s a statistic for
measuring the accuracy of machine translations com-
pared to human translators.
REFERENCES
Alomari, A., Idris, N., Sabri, A., and Alsmadi, I. (2021).
Deep reinforcement and transfer learning for abstrac-
tive text summarization: A review.
Chen, C., Zhang, R., Koh, E., Sungchul, Kim, S. C., Yu, T.,
Rossi, R., and Bunescu, R. (2019). Figure captioning
with reasoning and sequence-level training.
Guan, Y., Guo, S., Li, R., Li, X., and Zhang, H. (2021).
Frame semantics guided network for abstractive sen-
tence summarization.
Gupta, S. and Gupta, S. K. (2018). Abstractive summariza-
tion: An overview of the state of the art.
Hsu, T.-Y., Giles, C. L., and Huang, T.-H. K. (2021). Sci-
cap: Generating captions for scientific figures.
Huang, C.-Y., Hsu, T.-Y., Rossi, R., Nenkova, A., Kim,
S., Chan, G. Y.-Y., Koh, E., Giles, C. L., and Ting-
Hao’Kenneth’Huang (2023). Summaries as captions:
Generating figure captions for scientific documents
with automated text summarization.
James G. Mork, Antonio J. Jimeno Yepes, A. R. A. (2013).
The nlm medical text indexer system for indexing
biomedical literature.
Jing, B., Xie, P., and Xing, E. (2018). On the automatic
generation of medical imaging reports.
Khan, B., Shah, Z. A., Usman, M., Khan, I., and Niazi,
B. (2023). Exploring the landscape of automatic text
summarization: A comprehensive survey.
Li, C., Wong, C., Zhang, S., Usuyama, N., Liu, H., Yang,
J., Naumann, T., Poon, H., and Gao, J. (2024). Llava-
med: Training a large language-and-vision assistant
for biomedicine in one day.
Li, Y., Liang, X., Hu, Z., and Xing, E. (2018). Hybrid
retrieval-generation reinforced agent for medical im-
age report generation.
Lin, H. and Ng, V. (2019). Abstractive summarization: A
survey of the state of the art.
Liu, Y. (2019). Fine-tune bert for extractive summarization.
Mike Lewis and, Y. L., Goyal, N., Ghazvininejad, M., Mo-
hamed, A., Levy, O., Stoyanov, V., and Zettlemoyer,
L. (2020). Bart: Denoising sequence-to-sequence pre-
training for natural language generation, translation,
and comprehension.
Raffel, C., Shazeer, N., Roberts, A., Lee, K., Narang, S.,
Matena, M., Zhou, Y., Li, W., and Liu, P. J. (2019).
Exploring the limits of transfer learning with a unified
text-to-text transformer.
Shin, H., Roberts, K., Lu, L., Demner-Fushman, D., Yao,
J., and Summers, R. (2016). Learning to read chest
x-rays: recurrent neural cascade model for automated
image annotation.
Shukre, S., Salunkhe, S., Rathi, P., Shinde, V., and Mane,
P. M. V. (2023). Research paper summarization using
nlp.
Thompson, P. P.-S. J. M. C. S. H. (2017). Research: The
readability of scientific texts is decreasing over time.
Yuxuan Xiong, Bo Du, P. Y. (2019). Reinforced transformer
for medical image captioning.
Zachary, V. V., Trillo, J., Abalorio, C., Bustillo, J., Bojocan,
J., and Elape, M. (2022). Ocr-based hybrid image text
summarizer using luhn algorithm with finetunetrans-
former modelsfor long document.
Zhang, Z., Xie, Y., Xing, F., McGough, M., and Yang, L.
(2017). Mdnet: A semantically and visually inter-
pretable medical image diagnosis network.
Zhong, M., Liu, P., Chen, Y., Wang, D., Qiu, X., and Huang,
X. (2020). Extractive summarization as text matching.
A Multimodal Approach to Research Paper Summarization
957
