
Domain Ontology for Semantic Mediation in the Data Science Process
Silvia Lucia Borowicc
1 a
and Solange Nice Alves-Souza
2 b
1
School of Arts, Sciences and Humanities, Universidade de S
˜
ao Paulo, Arlindo Bettio 1000, S
˜
ao Paulo, Brazil
2
Polytechnic School, Universidade de S
˜
ao Paulo, S
˜
ao Paulo, Brazil
Keywords:
Semantic Mediation, Data Science, Data Integration, Ontology, Public Health.
Abstract:
The integration of heterogeneous data sources is a persistent challenge in public health. As regards dengue
and other arboviral diseases, data collected over many years by various organizations are fragmented across
heterogeneous, siloed databases, lacking semantically consistent integration for effective decision-making in
health crises. These organizations operate autonomously but must collaborate to integrate data for a unified
understanding of the domain of knowledge. However, standardizing infrastructure or unifying systems is often
unfeasible. This study proposes the incorporation of semantic mediation into the data science process, intro-
ducing an innovative approach for data mapping that preserves the autonomy of data providers while avoiding
interference with their existing infrastructure or systems. The goal is to streamline the integration and analysis
of distributed, heterogeneous datasets by applying domain ontologies within an iterative data science process.
Unlike traditional approaches, which perform data mapping in later stages, our approach advances this step to
the definition phase, providing benefits such as early standardization, greater efficiency and error reduction.
The methodology includes a collaborative workflow for constructing a modular domain ontology that will sup-
port the data mapping from data sources to a global RDF ontology-based data model. This approach fosters
expert involvement and accommodates evolving domain knowledge. The results demonstrate that semantic
mediation enables the consolidation of semantically enriched data, enhancing the understanding of outcomes
in the decision-making process, in a scalable process for integrating public health data in epidemiological
monitoring and response contexts.
1 INTRODUCTION
Health systems face challenges regarding data man-
agement and integration. The difficulty in addressing
questions with up-to-date information about the effec-
tiveness of preventive health policies and strategies,
operational costs, hospitalization rates, and mortality
cases highlights the technological and structural lim-
itations of the health systems. Frequently, the mere
existence of data is insufficient to generate the nec-
essary information for effective and timely decision-
making. The lack of integration between systems
and data heterogeneity restrict an efficient and coordi-
nated response, especially in crisis scenarios (Borow-
icc and Alves-Souza, 2024).
Addressing Dengue, a persistent endemic in re-
gions with established populations of the vector Aedes
aegypti, has led research institutions, and munici-
pal, state-level, and federal health agencies, to ac-
a
https://orcid.org/0000-0001-7399-274X
b
https://orcid.org/0000-0002-6112-3536
cumulate substantial volumes of data on disease in-
cidence, climatic conditions, infestation rates, and
other relevant variables over more than 20 years.
However, these datasets remain dispersed across vari-
ous research centers and institutions at different ad-
ministrative levels. They were neither integrated
nor originally designed with interoperability require-
ments. Additionally, they were developed at differ-
ent times, using diverse structures and technologies,
which poses significant challenges for data integra-
tion and cross-referencing within computational sys-
tems (Brasil, 2009).
In public health, whereby rapid and evidence-
based decisions are crucial, the lack of precision
and the inability to effectively integrate and utilize
these data presents a substantial barrier (Aquino et al.,
2023; Pinto et al., 2024). Data heterogeneity, with
distinct terminologies and formats, complicates inte-
gration and limits the use of information for analysis,
outbreak monitoring, and planning preventive and re-
sponse actions. The worsening epidemiological sce-
nario, marked by a record increase in dengue cases in
234
Borowicc, S. L. and Alves-Souza, S. N.
Domain Ontology for Semantic Mediation in the Data Science Process.
DOI: 10.5220/0013279600003929
In Proceedings of the 27th International Conference on Enterprise Information Systems (ICEIS 2025) - Volume 1, pages 234-242
ISBN: 978-989-758-749-8; ISSN: 2184-4992
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

Brazil in 2024, underscores the pressing demand for
insights that support new response strategies to com-
bat the disease and its vector (Brasil, 2024a).
Public policy management generally involves di-
verse stakeholders. In the Brazilian Unified Health
System (SUS), the management model is tripartite,
involving municipalities, state-level, and federal gov-
ernment entities. There is an interdependent flow of
data among information systems managed by the SUS
Informatics Department (DataSUS) and those main-
tained by states and municipalities, as well as data ex-
changes with governmental and civil society organi-
zations. Additionally, states and municipalities have
different demands and resources. These characteris-
tics describe a vast and complex context in which co-
operation is essential, as the entities involved operate
autonomously, with no oversight from one over the
others (Brasil, 2024b).
Therefore, the hypothesis of unifying systems, in-
tegrated software solutions, or standardizing infras-
tructure to serve all, as proposed in (Tour
´
e et al.,
2023), becomes unfeasible due to the significant dif-
ferences in individual requirements and resources,
with only partial overlaps. Nonetheless, the demand
for access to integrated data and information is global,
especially for health managers responsible for design-
ing, supervising, and evaluating public policies. Clear
understanding of the information is essential for all
stakeholders using integrated data. The complexity
of such integration is already evident, given the het-
erogeneity of the various municipal, state-level, and
DataSUS systems developed over time and with dif-
ferent technologies. Thus, the question arises: how
can data be integrated while respecting autonomy, lo-
cal and global requirements, and the premise that the
meaning of data and information is clear and accessi-
ble?
This article addresses semantic mediation in data
integration in the field of public health, with a specific
focus on data related to Dengue and other arboviruses.
We propose a data science process that incorporates
semantic mediation to integrate data from heteroge-
neous and independent sources. A domain ontology
focused on Dengue, describing the global data model,
was created to support the mapping of data sources
and interpretation of data analysis results. Ontolo-
gies provide a formal structure to represent a domain
of knowledge, enabling concepts and relationships
within the data to be consistently described.
The incorporation of semantic mediation into the
data science process aims to provide effective map-
ping between data sources and the global model,
while respecting the autonomy of the data-providing
entities.
The domain ontology, initially focused on
Dengue, should be flexible, designed to incorporate
new concepts and expand its scope to include other
arboviruses and diseases that impact the Health Sys-
tem and are the target of public health policies. This
ontology may be applied by applications and plat-
forms that require reliable information about the dis-
ease, or for modeling and inference mechanisms, as
well as to provide useful information to educate the
general population. There are several initiatives with
data and information about Dengue, such as (Brasil,
2024a), (Brasil, 2024c) and (Codeco et al., 2018), al-
though each one is isolated, maintaining its own dis-
tinct repository.
This approach will enable the use of a common
repository that can be collaboratively enhanced, be-
coming increasingly robust and reliable, provided
there is strict control over the method of information
update. Thus, the concepts described in the ontol-
ogy should be accessible and incrementally enhanced
in a collaborative manner. Therefore, this article
also proposes a process for the defining and develop-
ing the domain ontology, using a metadata repository
inspired by Wiki systems (Cunningham and Leuf,
2002). The process enables modular and collaborative
construction, with versioning implemented to docu-
ment and preserve the history of the knowledge bases
used to semantically describe the data.
2 CONCEPTS AND PRIOR
RESEARCH
Data availability faces challenges, as noted by
(Gasc
´
o-Hern
´
andez et al., 2018), including organiza-
tional culture, compliance with legislation and regu-
lations, lack of infrastructure and standards, data frag-
mentation, and legacy systems, which are outdated
and based on discontinued technologies. Financial
constraints and the lack of resources also impact the
ability of public agents to develop and to maintain ad-
equate solutions. The insufficient knowledge about
the data and metadata lifecycle is another issue that
hinders data management and usage. Another chal-
lenging aspect is that domain knowledge is not typ-
ically explicit in the datasets (Borowicc and Alves-
Souza, 2024).
Different organizations may store similar data in a
heterogeneous manner, making it difficult to merge
datasets effectively (Deshpande et al., 2023). Al-
though some public administration institutions pro-
vide open data, these data are fragmented, remaining
siloed, and are often associated with a set of metadata
that is insufficient for the community to properly un-
Domain Ontology for Semantic Mediation in the Data Science Process
235

derstand and use (Coeli, 2006; Carvalho et al., 2011;
Gasc
´
o-Hern
´
andez et al., 2018).
These issues are consistent with the challenges
identified by (Bassi and Alves-Souza, 2023), which
include the existence of data silos and the lack of
standardized metadata as critical barriers to imple-
menting effective data governance, directly impacting
decision-making.
The Swiss Personalized Health Network (SPHN)
project introduced a framework and a standardized
infrastructure across hospitals, under the responsi-
bility of centralized coordination. The project aims
to create a health research repository based on the
FAIR principles (Findable, Accessible, Interoperable,
Reusable) (Wilkinson, 2016), with common models
and formats. The authors do not detail the implemen-
tation of the model but describe the requirement for
the hospitals involved to adhere to the defined stan-
dards and deliver the data in a specific format (Tour
´
e
et al., 2023).
The hypothesis of using a single database through
a unified system does not apply to contexts in which
data is produced by different organizations that op-
erate autonomously in their management but are in-
terdependent for obtaining information and a global
view of the domain for analysis and decision-making.
Ontologies offer valuable contributions to data in-
tegration and harmonization, particularly as regards
heterogeneous data sources. Defined by classes, prop-
erties, and relationships, ontologies provide a formal
and structured representation of knowledge within a
specific domain to create a common conceptual model
(Wache et al., 2001; Mahmoud et al., 2021).
From the Semantic Web field, the Resource De-
scription Framework (RDF)
1
is the standard model
for information representation, consisting of a graph
that formally describes the semantics or meaning of
the information. Heterogeneous data sources for an
RDF model can be mapped by the expression of rules
using the RDF Mapping Language (RML
2
). These
models and languages are open standards, thus com-
plying with public data management and also with the
FAIR principles.
In this work, the requirement for autonomy
of municipal and state-level health administrations
makes the standardization of infrastructure unfeasi-
ble. Therefore, the inclusion of semantic mediation
based on ontologies and Semantic Web technologies
is proposed to address data heterogeneity during the
data science process.
1
http://www.w3.org/RDF/
2
https://rml.io/specs/rml/
3 SEMANTIC MEDIATION IN
THE DATA SCIENCE PROCESS
The data science process is described by (Dekht-
yar, 2023) as a sequence of steps involving problem
identification, data collection, preparation, modeling,
analysis, and communication of results, in an iter-
ative process. Other documents that also describe
the data science lifecycle, such as (Boenig-Liptsin
et al., 2022), (Stodden, 2020), and (Keller et al.,
2020), complement this view by applying the se-
quence to specific contexts and incorporating dimen-
sions of ethics or governance. Although there are
mentions, as in (Borgman, 2019), of the importance
of the contextual interpretation of data, semantic ap-
proaches are not explored. Data mapping tasks are
traditionally performed after data collection, during
the treatment or preprocessing stages (Kimball and
Caserta, 2004).
In contrast, this article proposes the introduction
of semantic mediation steps that precede data collec-
tion and guide the integration of heterogeneous data.
The mediation is based on ontologies and aims to pro-
vide effective mapping between data sources and the
global model, without interfering with the manage-
ment of the data providers, ensuring that the data can
be integrated and made available to all stakeholders
(Borowicc and Alves-Souza, 2024).
As of the progress of this research, no proposal
or solution has been found that integrates data on
Dengue, or other urban arboviruses, from munici-
pal, state-level, and SUS information systems in a
way that allows for analyzing information and actions
taken for disease control by different initiatives over
more than 20 years (Brasil, 2009).
The proposal intends to enhance the integration
and analysis process by ensuring that the data, despite
coming from heterogeneous sources, are interpreted
and used in a consistent and consolidated manner.
Defining a semantic structure from the early stages
of the process enables a precise alignment with the
problem to be analyzed, ensuring that only relevant
data is collected and integrated. Furthermore, deci-
sions on which data to collect are based on clear cri-
teria, respecting the autonomy of the data providers
and compliance with regulations in public data man-
agement. This approach also enhances data under-
standing and querying, providing a solid foundation
for reliable analysis and contextualized results.
The proposed data science process with semantic
mediation is structured iteratively, as in the original
cycle (Dekhtyar, 2023), in which questions and hy-
potheses are formulated based on the results. How-
ever, it allows for refining and extending the domain
ICEIS 2025 - 27th International Conference on Enterprise Information Systems
236

ontology when a new iteration of the process is ini-
tiated. As shown in Figure 1, this cycle is supported
by semantic enrichment based on the definition of a
domain ontology, and the mapping of data sources to
the data model created from it. Semantic mediation is
instrumental in structuring, integrating heterogeneous
data, and in interpreting and using the resulting anal-
ysis.
Figure 1: Data science process with semantic mediation.
The steps and the role of semantic mediation in
the process is described as follows.
• 1. Formulation of Questions. The process be-
gins with the definition of the research questions
and the specific objectives of the analysis. In this
step, the areas of interest are identified to guide
the definition or the extension of the domain on-
tology.
• 2. Domain Ontology Definition. To ensure
a common understanding among different data
sources, it is necessary to define, construct, or ex-
tend an ontology that represents the domain of the
analysis. The ontology organizes knowledge in a
structured manner, contributing to the definition
of scope, global conceptual model, and the priori-
tization of data collection and processing.
• 3. Semantic Mapping of Sources. In this step,
the semantic mapping of data sources is per-
formed to ensure that each term and concept is
correctly associated with the domain ontology.
This enables the harmonization of different termi-
nologies and facilitates the integration process.
• 4. Data Collection. With the sources seman-
tically mapped, the data collection process is
optimized. In this phase, relevant data is acquired
from data sources, such as databases, infor-
mation systems, and structured files.
• 5. Preprocessing. The collected data are pro-
cessed for cleaning, removing inconsistencies,
and other activities to ensure data quality.
• 6. Analytical Modeling. In this step, the prepro-
cessed data is organized and structured to ensure
its suitability for analysis. Statistical techniques
and machine learning algorithms are applied to
prepare the data by selecting and extracting rele-
vant features. The analytical modeling step is en-
hanced by using semantically mapped and well-
organized data, which facilitates the accurate in-
terpretation of results.
• 7. Data Analysis. After preparing the data in
the analytical modeling phase, the model is used
to analyze the data, identifying patterns, clus-
ters, and performing classifications. Data analy-
sis aims to generate metrics and insights that ad-
dress the research questions formulated at the out-
set. Semantic mediation contributes to ensuring
that the results are consistently contextualized and
aligned with the research objectives.
• 8. Results Presentation. The results of the anal-
ysis are presented for interpretation. Visualiza-
tion tools help communicate results and display
information in an accessible and informative way,
allowing users and analysts to draw meaningful
conclusions.
• 9. Analysis of Results. The results are evaluated
based on the initial questions. This step allows for
assessing the quality of the findings and identify-
ing areas in which the analysis or methodology
may need refinement.
• 10. Problem Refinement and Follow-up. Based
on the analysis of results, the initial objectives
may be refined. The process may return to the
question formulation stage, creating an iterative
cycle in the data science process.
In contexts of autonomy and interdependence
between data-providing and data-consuming institu-
tions, whereby multiple data sources need to be inte-
grated in a collaborative effort, performing the map-
ping between the sources and the global model at the
definition stage is a practice that can enhance effi-
ciency and improve data quality and, consequently,
the results of the analyses (Borowicc and Alves-
Souza, 2024).
Domain Ontology for Semantic Mediation in the Data Science Process
237
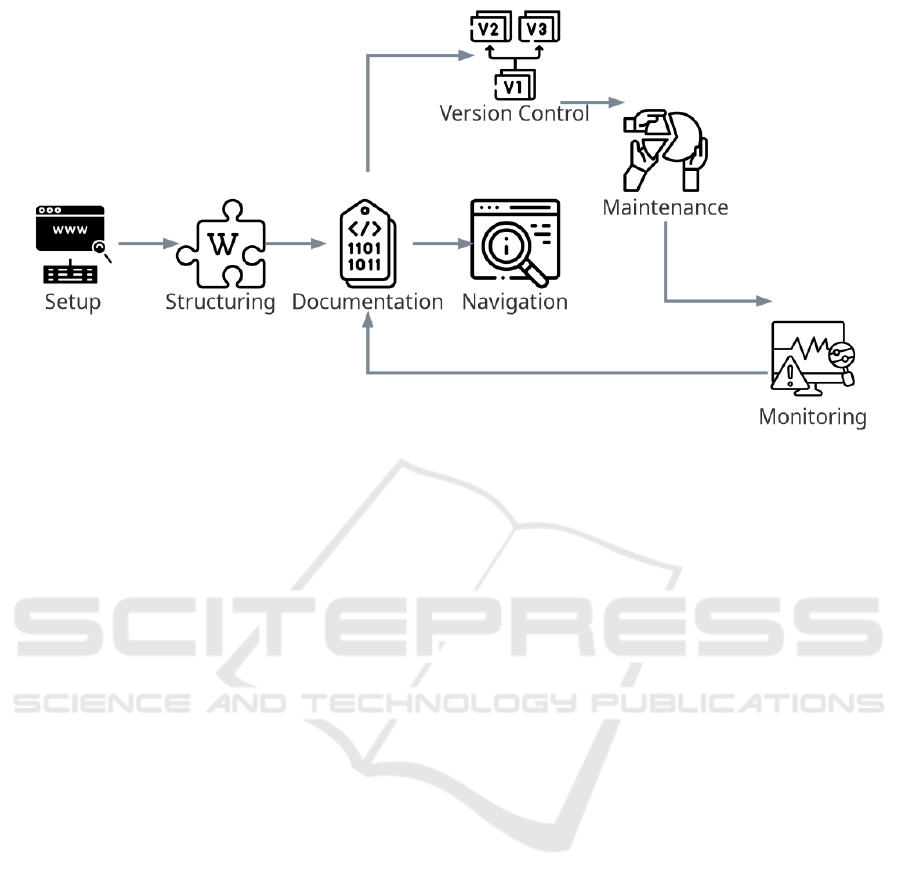
Figure 2: Metadata repository construction and maintenance workflow.
3.1 Methodology for Domain Ontology
Construction
Although the use of ontologies is referenced in
previous works on data integration, the process of
their construction remains under-discussed. However,
this is a complex process that encompasses multiple
stages for gathering reliable information, which ulti-
mately contributes to the knowledge needed for on-
tology construction. Therefore, this section outlines
the methodology employed in the research presented
herein.
The initial activities to understand the researched
domain involved meetings with experts in arbovirus
surveillance and control, as well as with healthcare
service teams. These discussions were crucial for
gaining a comprehensive understanding of the do-
main, helping to create an overview of the terms used
in operational activities. Many of these terms are par-
tially reflected in the existing information systems and
are a part of the data sources.
A research conducted by (Lazarre et al., 2022) dis-
cusses various techniques and approaches considered
state-of-the-art in the process of ontology creation and
management. Inspired by this study, an iterative cy-
cle for creating and maintaining a metadata reposi-
tory was proposed, considering relevant requirements
in ontology development, such as:
• Collaboration Platform. Facilitates the
contribution of team members, fostering a collab-
orative environment, thus enhancing the quality
and scope of the knowledge representation.
• Flexibility and Modularity. A modular approach
increases flexibility and enables quick and effi-
cient updating and expansion of documentation.
• Accessibility. A repository provides a shared un-
derstanding of the domain knowledge, and its in-
formation can be accessed and used by users from
diverse interest groups.
• Review and Validation. Periodic reviews by ex-
perts ensure that definitions and information re-
main consistent and useful for different user pro-
files.
• Transparency. The platform can maintain a his-
tory of changes and contributions, offering a clear
view of modifications made to the metadata.
These elements enhance the design and manage-
ment of ontologies in a collaborative environment
(Lazarre et al., 2022). The proposed metadata reposi-
tory is an implementation inspired by the Wiki sys-
tems model from (Cunningham and Leuf, 2002),
which leads the resources to meet the mentioned re-
quirements. The workflow for creating and maintain-
ing the metadata repository is shown in Figure 2 and
detailed below.
The workflow begins with the setup task, during
which the appropriate infrastructure is selected and
configured to store metadata. It then moves to the
structuring task, whereby pages and categories are
created to organize the content. In the documenta-
tion task, essential content regarding concept descrip-
tions, data providers, data sources, transformations,
and processing rules are recorded.
ICEIS 2025 - 27th International Conference on Enterprise Information Systems
238
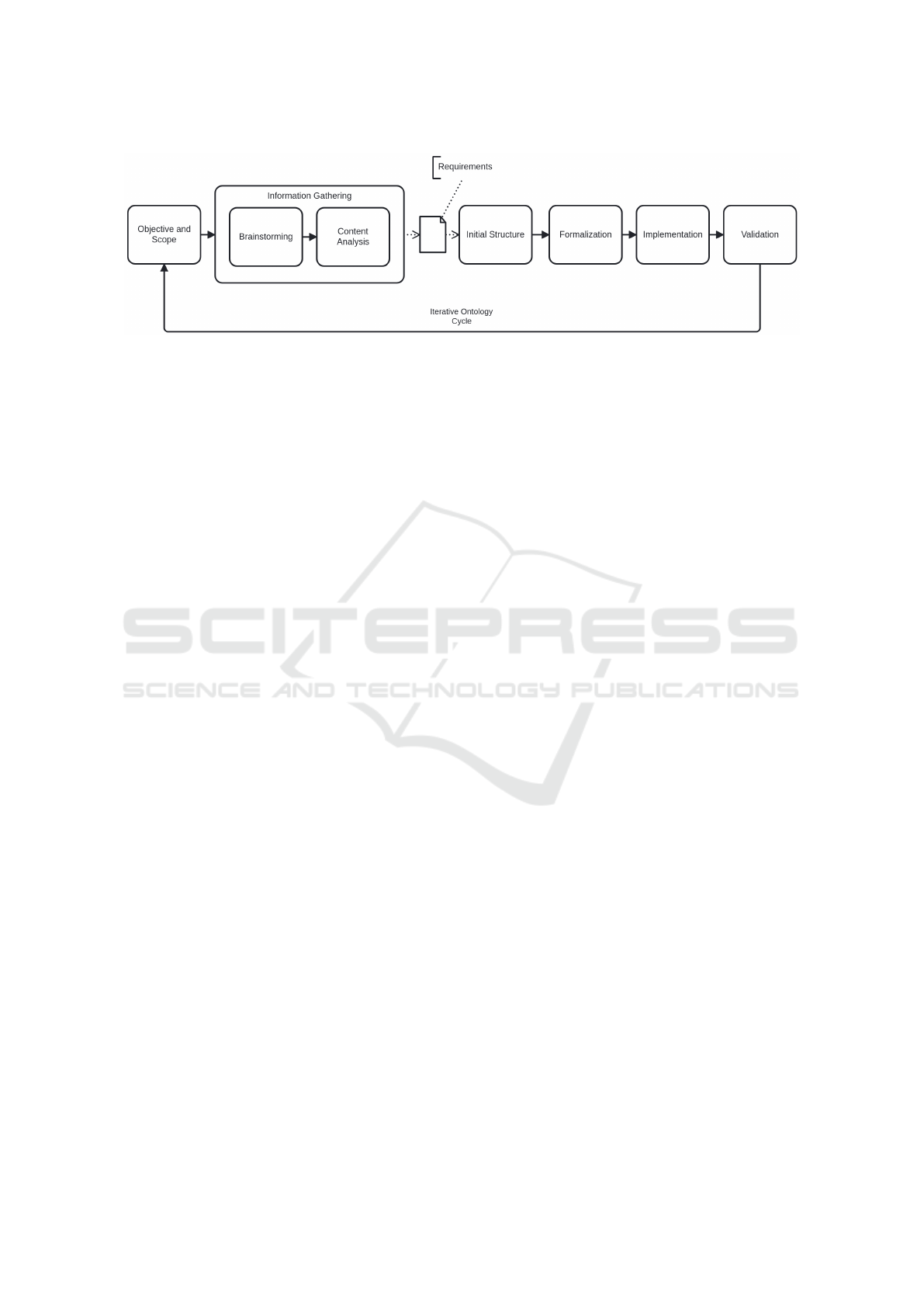
Figure 3: Domain Ontology Construction Process.
After documentation, navigation is implemented
to provide access to the metadata, along with version
control to track changes. Collaborative monitoring
and maintenance ensure that the documentation re-
mains updated and enable continuous improvements,
with updates feeding back into the documentation.
In the proposed data science process, the ontology
definition can be achieved by selecting and extending
existing ontologies, or by creating a domain-specific
ontology. To the best of our knowledge, there are
no Portuguese-language ontologies that describe the
knowledge domain of Dengue surveillance and con-
trol, or other arboviral diseases. Although ontologies
are suggested for use in data integration projects, pri-
marily to address data heterogeneity, their construc-
tion within a data science process remains underex-
plored (Borowicc and Alves-Souza, 2024). We thus
propose an iterative process, shown in Figure 3, for
defining the domain ontology to support semantic me-
diation in the proposed data science process.
The process starts by defining an initial scope,
which is associated to the questions formulated dur-
ing the data science process - in this context, the scope
covered activities related to epidemiological and ento-
mological surveillance, as well as the monitoring of
reported clinical cases. Once the scope was estab-
lished, the process of gathering domain-specific in-
formation began, which included: (I) brainstorming:
discussions with experts to understand the workflows
involved in Dengue and arboviral disease surveillance
and control operations, as well as the terminology
used by the stakeholders; (II) content analysis: re-
viewing documents related to regulations and guide-
lines that regulate the activities, analyzing data mod-
els, examining application source code, and studying
forms used for data collection.
The experts provided valuable insights into the op-
eration of the Dengue surveillance and control pro-
gram, highlighting the challenges faced in using the
data collected for decision-making and in ensuring the
accessibility of public data. Additionally, they helped
identify the technical terminology used in reports and
operational documents, leading to the creation of an
initial glossary that facilitated the understanding of
data. The system and documents contained acronyms
and reported field names that lacked adequate descrip-
tions or had insufficient explanations for proper data
interpretation. Throughout the course of the project,
this glossary was integrated into the metadata reposi-
tory.
Reviewing documents, such as those outlining the
guidelines for the Brazilian National Dengue Control
Program (PNCD) (Brasil, 2009) and other public doc-
uments provided by municipal and state authorities,
was essential for understanding the surveillance and
control activities. These documents provided techni-
cal guidelines and standards that enriched the under-
standing of the knowledge domain.
We conducted a detailed analysis of the data
model generated through reverse engineering of the
databases from the information systems currently in
use. This analysis was essential for understanding the
structure of the data collected by the information sys-
tems employed in the operations. The key data mod-
els analyzed, along with the associated documents,
are part of the Aedes Surveillance and Control Sys-
tem (SISAWEB) and the Notification of Diseases In-
formation System (SINAN), including:
• SISAWEB - Visit Records. Contains informa-
tion on field visits, including dates, locations, and
inspection outcomes. This database model repre-
sents records of visits conducted by surveillance
and health agents to specific properties or areas,
documenting the conditions observed and the in-
terventions implemented.
• SISAWEB - Territorial Division. Describes the
territorial division structure used in the surveil-
lance system, outlining the spatial organization of
the data. This division is essential for understand-
ing how the data is geographically segmented and
how different regions are monitored.
• SINAN - Case Notification Records. Contains
data on case notifications, including detailed epi-
Domain Ontology for Semantic Mediation in the Data Science Process
239
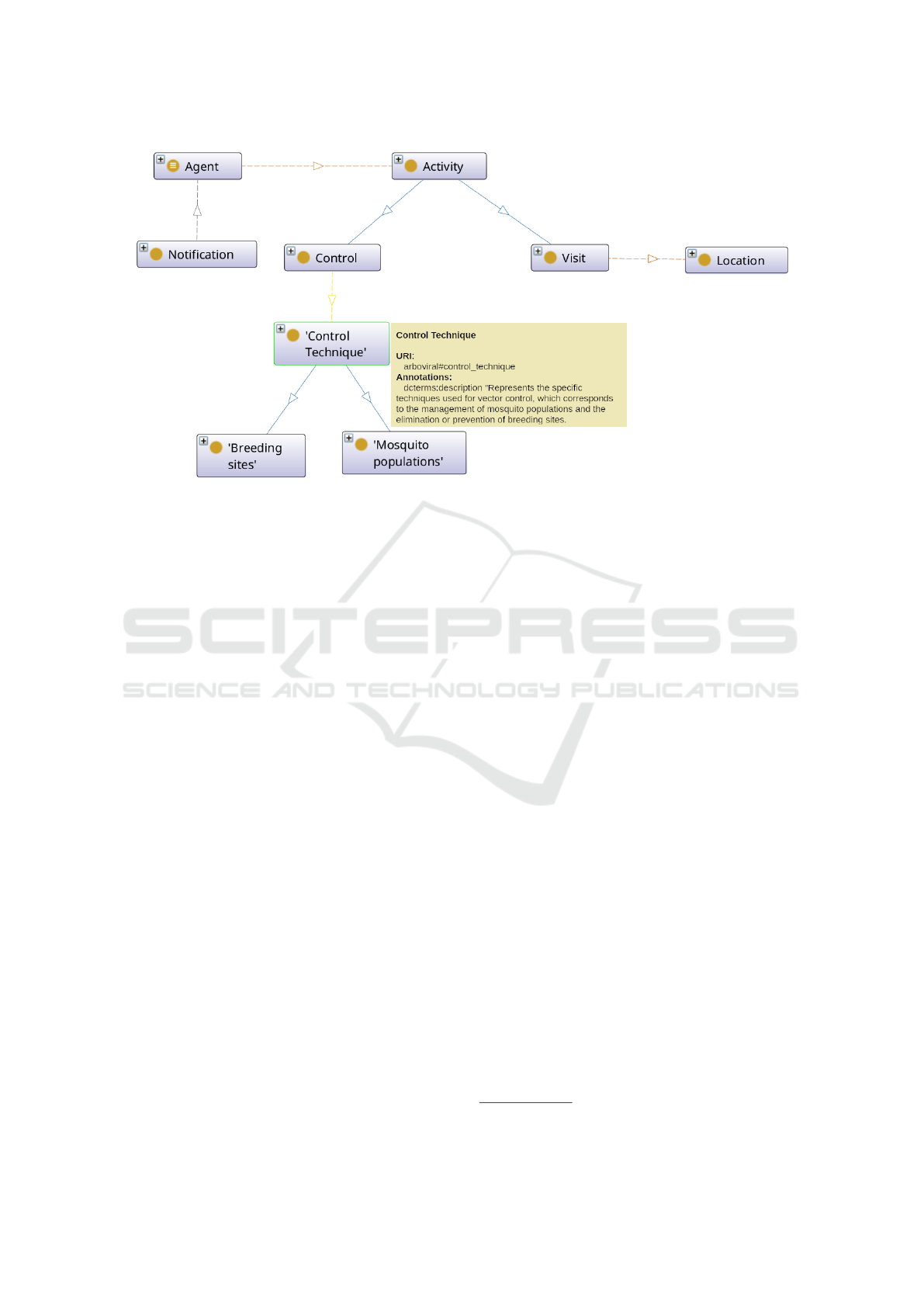
Figure 4: Fragment of the Dengue domain ontology.
demiological information. This database is vital
for tracking the incidence of arboviral cases and
for analyzing outbreak trends over time.
Besides analyzing the data models, we also re-
viewed the source code of the SISAWEB system.
This analysis uncovered additional descriptions and
classifications that were not present in the data model.
The source code provided valuable insights into the
operational logic and data processing rules. Among
the key findings were rules implemented within the
code that determined how specific data were classified
and processed. For example, the code contained logic
for distinguishing between suspected and confirmed
cases, as well as for grouping data by time periods.
Additionally, we examined conditional structures that
influenced how data were recorded and displayed in
reports. These structures were instrumental in under-
standing the behavior of the system in generating re-
ports and conducting analyses.
Furthermore, to gain insight into the data accessed
by the application during report generation, we ana-
lyzed the database logs. This step was essential for
understanding the data structure. For example, infor-
mation on possible larval habitats was encoded in ta-
bles labeled 1 and 2, with no associated descriptions
explaining the numbering scheme. Only by analyzing
the logs and the system could we identify that Table
2 corresponded to the possible larval habitats listed in
the visit record table, while value 1 referred to records
of other field activities.
The SINAN disease notification form was used to
identify the various entities present in the data files
provided by the Brazilian Ministry of Health (MS).
This analysis supplemented the information obtained
in earlier stages, providing a more thorough under-
standing of the data structures employed.
The SINAN notification forms are standardized
documents used to record cases of compulsory-
notification diseases, including Dengue. These forms
include specific fields for capturing patient personal
information, clinical and epidemiological data, as
well as follow-up details. This information is essen-
tial for epidemiological surveillance and plays a crit-
ical role in monitoring and controlling disease out-
breaks. The key data included in the form are as fol-
lows:
• Personal Data. Name, age, gender, address.
• Clinical Data. Date of symptom onset, signs and
symptoms presented.
• Follow-up Data. Laboratory test results, case
progression and outcome.
3.2 Ontology Formalization
Based on the information gathered in the previous
stages, we proceeded with the creation of the Dengue
domain ontology using the free, open-source ontol-
ogy editor Prot
´
eg
´
e
3
. The classes identified were de-
scribed in the ontology, which is partially presented
in Figure 4. This ontology serves as the global data
model on which semantic mediation is based. It de-
fines key classes, such as:
• Visit. Represents a field visit, with attributes such
as date, location, and outcome.
3
https://protege.stanford.edu/
ICEIS 2025 - 27th International Conference on Enterprise Information Systems
240

• Location. The territorial structure used in the
surveillance and control system.
• Control Technique. Represents the specific tech-
niques used for vector control, which corresponds
to the management of mosquito populations and
the elimination or prevention of breeding sites.
• Notification. Information on notifications of
Dengue and other arboviral cases.
Besides the information specific to surveillance
and control programs, as well as disease incidence
data, references to scientific literature were incorpo-
rated into the described ontology to further enrich it
with supplementary information, thereby strengthen-
ing its semantic layer. Articles and publications on
Dengue and Aedes aegypti control, indexed in sci-
entific databases such as PubMed and SciELO, were
used in this process.
Integration with scientific databases allows asso-
ciating epidemiological and surveillance data with
relevant scientific literature, facilitating research and
decision-making. For this, the ontology was extended
to include classes and properties representing scien-
tific articles, authors, journals, and keywords.
Thus, the ontology enabled the structured and se-
mantic modeling of the information relevant to the
research domain, serving as a key component of the
proposed semantic mediation within the data science
process.
For effective data integration, mapping between
data sources and the global model is essential. As
highlighted in the literature as the state-of-the-art for
semantic annotation of structured data, the Karma
tool performs mapping by loading a dataset or a sub-
set of data from the source (Borowicc and Alves-
Souza, 2024). In contrast, the proposed semantic me-
diation approach enables mapping directly from the
data dictionary that describes the source, eliminating
the need to retrieve data prior to this step or to load
data for processing the task.
4 CONCLUSIONS
This study explored a semantic mediation approach
incorporated into the data science process, focusing
on the integration of heterogeneous public health data,
particularly for Dengue and other arboviral diseases.
The creation of a domain ontology was a key step,
providing a structured semantic framework for data
integration and analysis. The ontology was developed
to represent knowledge on Dengue surveillance and
control, and is designed to be modular and expandable
as new data sources are incorporated.
By introducing a structured workflow for the col-
laborative development of the domain ontology, the
approach enhances the integrated analysis of hetero-
geneous data sources. It ensures consistency from the
outset and facilitates expert collaboration, allowing
continuous updates to the semantic model. Seman-
tic mediation plays a central role in ensuring that data
is consistently interpreted and integrated, regardless
of its origin.
The use of a data dictionary for mapping local data
sources to the global model will be discussed in a fu-
ture paper, focusing on process automation to opti-
mize data integration processes.
The key contributions of this work include an-
ticipating semantic mapping in the definition phase,
establishing a consistent and standardized data foun-
dation early on. This approach reduces rework dur-
ing data transformation, improving efficiency and the
quality of analyses. Additionally, the modular and
collaborative metadata repository enhances the ontol-
ogy flexibility, supporting continuous expansion.
While the approach shows significant advance-
ments, challenges remain in automating the seman-
tic mapping process and expanding the ontology to
multiple domains. Future work should address issues
such as ontology governance, maintaining semantic
consistency at scale, and managing complex models.
The continuation of this work has the potential
to significantly improve public health data manage-
ment systems, supporting decision-making and en-
abling faster and coordinated responses to complex
epidemiological challenges.
ACKNOWLEDGEMENTS
Authors are grateful for the support given by
S
˜
ao Paulo Research Foundation (FAPESP). Grant
#2023/10080-3.
REFERENCES
Aquino, E., Borowicc, S., Alves-Souza, S., Teixeira, R.,
Ishitani, L., Malta, D., and Morais Neto, O. (2023).
Distribution of garbage codes in the mortality infor-
mation system, brazil, 2000 to 2020. Cien Saude Co-
let. In press.
Bassi, C. A. and Alves-Souza, S. N. (2023). Challenges to
implementing effective data governance: A literature
review. In Proceedings of the 15th International Joint
Conference on Knowledge Discovery, Knowledge
Engineering and Knowledge Management (IC3K
2023) - Volume 3: KMIS, pages 17–28, Lisbon,
Portugal. SCITEPRESS – Science and Technology
Domain Ontology for Semantic Mediation in the Data Science Process
241

Publications, Lda.
Boenig-Liptsin, M., Tanweer, A., and Edmundson, A.
(2022). Data science ethos lifecycle: Interplay of
ethical thinking and data science practice. Journal
of Statistics and Data Science Education, 30(3):228–
240.
Borgman, C. L. (2019). The lives and afterlives of data.
Harvard Data Science Review, 1(1).
Borowicc, S. and Alves-Souza, S. (2024). Heterogeneous
Data Integration: A Literature Scope Review:. In Pro-
ceedings of the 26th International Conference on En-
terprise Information Systems, pages 189–200, Angers,
France. SCITEPRESS - Science and Technology Pub-
lications.
Brasil, M. d. S. d. B. (2024a). Painel de monitoramento das
arboviroses. Acesso em: 8 nov. 2024.
Brasil, M. d. S. M. (2009). Diretrizes nacionais para a
prevenc¸
˜
ao e controle de epidemias de dengue. S
´
erie
A. Normas e Manuais T
´
ecnicos. Minist
´
erio da Sa
´
ude,
Bras
´
ılia.
Brasil, M. d. S. M. (2024b). Comiss
˜
ao intergestores tripar-
tite. Acesso em: 8 nov. 2024.
Brasil, M. d. S. M. (2024c). Doenc¸as e
agravos de notificac¸
˜
ao de 2007 em diante
(sinan). https://datasus.saude.gov.br/acesso-a-
informacao/doencas-e-agravos-de-notificacao-de-
2007-em-diante-sinan/. Accessed: 2024-11-17.
Carvalho, C. N., Dourado, I., and Bierrenbach, A. L. (2011).
Subnotificac¸
˜
ao da comorbidade tuberculose e aids:
uma aplicac¸
˜
ao do m
´
etodo de linkage. Revista de
Sa
´
ude P
´
ublica, 45:548–555. Publisher: Faculdade de
Sa
´
ude P
´
ublica da Universidade de S
˜
ao Paulo.
Codeco, C., Coelho, F., Cruz, O., Oliveira, S., Castro, T.,
and Bastos, L. (2018). Infodengue: A nowcasting sys-
tem for the surveillance of arboviruses in brazil. Revue
d’
´
Epid
´
emiologie et de Sant
´
e Publique, 66:S386. Eu-
ropean Congress of Epidemiology “Crises, epidemio-
logical transitions and the role of epidemiologists”.
Coeli, C. M. (2006). Relacionamento de Bases de Dados
em Sa
´
ude. CADERNOS SA
´
UDE COLETIVA, 14(2).
Cunningham, W. and Leuf, B. (2002). The Wiki Way: Quick
Collaboration on the Web. Addison-Wesley, Boston,
MA.
Dekhtyar, A. (2023). DATA 301: Introduction to Data Sci-
ence. Lecture notes.
Deshpande, P., Rasin, A., Tchoua, R., Furst, J., Raicu,
D., Schinkel, M., Trivedi, H., and Antani, S. (2023).
Biomedical heterogeneous data categorization and
schema mapping toward data integration. Frontiers
in Big Data, 6. Publisher: Frontiers.
Gasc
´
o-Hern
´
andez, M., Martin, E. G., Reggi, L., Pyo, S., and
Luna-Reyes, L. F. (2018). Promoting the use of open
government data: Cases of training and engagement.
Government Information Quarterly, 35:233–242.
Keller, S. A., Shipp, S. S., Schroeder, A. D., and Korkmaz,
G. (2020). Doing data science: A framework and case
study. Harvard Data Science Review.
Kimball, R. and Caserta, J. (2004). The Data Warehouse
ETL Toolkit: Practical Techniques for Extracting,
Cleaning, Conforming, and Delivering Data. Wiley,
Indianapolis, IN.
Lazarre, W., Guidedi, K., Amaria, S., and Kolyang (2022).
Modular Ontology Design: A State-of-Art of Dis-
eases Ontology Modeling and Possible Issue. Revue
d’Intelligence Artificielle, 36(3):497–501.
Mahmoud, A., Shams, M. Y., Elzeki, O. M., and Awad,
N. A. (2021). Using semantic web technologies to
improve the extract transform load model. Comput-
ers, Materials & Continua, 68(2):2711–2726.
Pinto, L. F., Carvalho, A. A. d., and Pisco, L. A. C. (2024).
Inovac¸
˜
oes na gest
˜
ao da atenc¸
˜
ao prim
´
aria
`
a sa
´
ude,
contribuic¸
˜
oes dos inqu
´
eritos domiciliares e do censo
demogr
´
afico ibge (2022). Ci
ˆ
encia & Sa
´
ude Coletiva,
29(11):e07762024.
Stodden, V. (2020). The data science life cycle: A disci-
plined approach to advancing data science as a sci-
ence. Communications of the ACM, 63(7):58–61.
Tour
´
e, V., Krauss, P., Gnodtke, K., Buchhorn, J., Unni, D.,
Horki, P., Raisaro, J., Kalt, K., Teixeira, D., Crameri,
K., and
¨
Osterle, S. (2023). FAIRification of health-
related data using semantic web technologies in the
Swiss Personalized Health Network. Scientific Data,
10(1).
Wache, H., V
¨
ogele, T., Visser, U., Stuckenschmidt, H.,
Schuster, G., Neumann, H., and H
¨
ubner, S. (2001).
Ontology-based integration of information - a survey
of existing approaches. In Proceedings of the IJCAI-
01 Workshop on Ontologies and Information Sharing,
pages 108–118. CEUR Workshop Proceedings.
Wilkinson, M. D. (2016). Comment: The fair guiding prin-
ciples for scientific data management and stewardship.
Nature Publishing Group.
ICEIS 2025 - 27th International Conference on Enterprise Information Systems
242
