
Union and Intersection K-Fold Feature Selection
Artur J. Ferreira
1,3 a
and M
´
ario A. T. Figueiredo
2,3 b
1
ISEL, Instituto Superior de Engenharia de Lisboa, Instituto Polit
´
ecnico de Lisboa, Portugal
2
IST, Instituto Superior T
´
ecnico, Universidade de Lisboa, Portugal
3
Instituto de Telecomunicac¸
˜
oes, Lisboa, Portugal
fi
Keywords:
Explainability, Feature Selection, Filter, Interpretability, Intersection of Filters, K-Fold Feature Selection,
Union of Filters.
Abstract:
Feature selection (FS) is a vast research topic with many techniques proposed over the years. FS techniques
may bring many benefits to machine learning algorithms. The combination of FS techniques usually improves
the results as compared to the use of one single technique. Recently, the concepts of explainability and in-
terpretability have been proposed in the explainable artificial intelligence (XAI) framework. The recently
proposed k-fold feature selection (KFFS) algorithm provides dimensionality reduction and simultaneously
yields an output suitable for explainability purposes. In this paper, we extend the KFFS algorithm by perform-
ing union and intersection of the individual feature subspaces of two and three feature selection filters. Our
experiments performed on 20 datasets show that the union of the feature subsets typically attains better results
than the use of individual filters. The intersection also attains adequate results, yielding human manageable
(e.g., small) subsets of features, allowing for explainability and interpretability on medical domain data.
1 INTRODUCTION
The machine learning (ML) field is focused on learn-
ing from examples on a given dataset. The perfor-
mance of ML techniques can be improved by reduc-
ing the dimensionality of the input data by keeping
only the most relevant features, the key benefits are
faster training and better generalization performance.
For dimensionality reduction, the use of feature
selection (FS) techniques has been found appropri-
ate. FS aims to identify the best performing set of
features on a given task (Guyon et al., 2006; Guyon
and Elisseeff, 2003; Bolon-Canedo et al., 2015). FS
has a long research history and work towards improv-
ing FS techniques still continues (Alipoor et al., 2022;
Chamlal et al., 2022; Huynh-Cam et al., 2022; Jeon
and Hwang, 2023; Xu et al., 2022). FS techniques
can be grouped into four categories: filters, wrappers,
embedded, and hybrid (Guyon et al., 2006; Bolon-
Canedo et al., 2015). In this paper, we use filter tech-
niques, which assess the quality of subsets of features
by using some metrics over the data, without resorting
to any learning algorithm. In this sense, filter tech-
a
https://orcid.org/0000-0002-6508-0932
b
https://orcid.org/0000-0002-0970-7745
niques are referred to as agnostic. When dealing with
high-dimensional data, we often find that filters are
the only suitable approach, since the other techniques
are too time-consuming and their use becomes com-
putationally prohibitive (Hastie et al., 2009; Guyon
et al., 2006; Escolano et al., 2009). For recent
surveys on FS techniques, please see the publica-
tions by Remeseiro and Bolon-Canedo (2019), Pud-
jihartono et al. (2022a), and Dhal and Azad (2022).
In this work, we address the use of unsupervised
and supervised FS filter techniques for different types
of data. We propose to improve and extend the k-fold
feature selection (KFFS) algorithm proposed by Fer-
reira and Figueiredo (2023), using combinations of
heterogeneous filters. These combinations attain both
adequate dimensionality reduction and improved per-
formance. Moreover, the small dimensionality of re-
duced feature subspace allows for the human end user
to focus on explainability and interpretability tasks.
1.1 Combination of Filters
We find the use of combination of filters in different
applications. The problem of sleep disease diagnos-
tic was addressed by
´
Alvarez Est
´
evez et al. (2011),
with the monitoring of bio-physiological signals of
360
Ferreira, A. J. and Figueiredo, M. A. T.
Union and Intersection K-Fold Feature Selection.
DOI: 10.5220/0013282300003905
In Proceedings of the 14th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2025), pages 360-367
ISBN: 978-989-758-730-6; ISSN: 2184-4313
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

patients during sleep, with polysomnography (PSG)
data. A dataset with PSG of patients was used for
the detection of arousals in sleep. From a set of 42
features extracted from biosignals methods to detect
sleep events were developed. Using FS techniques
the goal was to remove redundant features, identify-
ing the best subset of features preserving classifica-
tion accuracy. Wrapper and filter methods and com-
binations of these were considered, by union and in-
tersection operations. Discarding the irrelevant fea-
tures, a reduced dimensionality dataset was obtained,
improving the accuracy of the classifiers.
The heterogeneous ensemble feature selection
(HEFS) method proposed by Damtew et al. (2023)
fuses the output feature subsets of five FS filters with
an union combination. It resorts to a merit-based eval-
uation to minimize redundancy of the obtained en-
semble of features. In a multi-class intrusion detec-
tion dataset, HEFS leads to better performance than
the individual FS methods.
Mochammad et al. (2022) proposed the multi-
filter clustering fusion (MFCF) technique. A multi-
filter method combining filter methods is applied as
a first step for feature clustering; then, the key fea-
tures are selected. The union of key features is used to
find all potentially important features. An exhaustive
search finds the best combination of selected features,
to maximize the accuracy of the classification model.
For rotating machinery problems, the fault classifica-
tion models using MFCF yields good accuracy.
The intersection of common features selected by
filter, wrapper, and embedded FS techniques was pro-
posed by Bashir et al. (2022). A support vector ma-
chines (SVM) classifier is then trained on medical do-
main data, attaining better results as compared to the
individual use of the FS methods.
Arya and Gupta (2023) introduced an ensemble
filter-based FS approach combining ANOVA, Pear-
son correlation coefficient, mutual information, and
Chi-square. The reduced feature sets are obtained
with the union and intersection operations. Using de-
cision tree, random forest, XGBoost, and CatBoost
classifiers on the Edge-IIoT dataset (cyber-attack de-
tection), we have 97.84% and 99.61% accuracy using
the intersection and union feature sets, respectively.
An ensemble FS approach was proposed by Seijo-
Pardo et al. (2017). The heterogeneous ensemble
combines the result of different FS methods, with the
same training data. The outputs of the base selec-
tors are combined with different aggregators to ob-
tain the resulting subset. On the experimental eval-
uation with the SVM classifier, ensemble results for
seven datasets achieve comparable on better perfor-
mance than the one attained by individual methods.
For reviews on ensemble FS methods and their
combination, please see the publications by Bol
´
on-
Canedo and Alonso-Betanzos (2019) and Pudji-
hartono et al. (2022b).
1.2 Paper Organization
The remainder of this paper is organized as follows.
In Section 2, we review some topics on feature selec-
tion. The proposed approach is detailed in Section 3.
The experimental evaluation is reported in Section 4.
Finally, Section 5 provides concluding remarks and
directions of future work.
2 FEATURE SELECTION
We introduce notation and review some details of FS
techniques in Section 2.1. An overview of the tech-
niques considered in this work is presented in Sec-
tion 2.2, including the k-fold feature selection (KFFS)
algorithm, which we propose to extend in this work.
2.1 Notation
Regarding the notation followed in this paper, let X =
{x
1
,... ,x
n
} denote a dataset, represented as a n × d
matrix (n instances on the rows and d features on the
columns). Each instance x
i
is a d-dimensional vector,
with i ∈ {1,. .. ,n}. Each feature vector, a column of
X, is denoted as X
j
, with j ∈ {1,...,d}. The num-
ber of classes is C, with c
i
∈ {1,...,C} represent-
ing the class of instance x
i
. Finally, y = {c
1
,. .. ,c
n
}
represents the class labels for each instance, with
c
i
∈ {1,. .. ,C}.
In this work, we consider both unsupervised and
supervised FS filters. The former do not use the class
label vector y, while the latter uses the label of each
instance to perform the feature assessment. Some FS
methods are based purely on the relevance of the fea-
tures; they rank the features according to some cri-
terion and then select the top-ranked ones. Other
methods are based on the relevance-redundancy (RR)
framework (Yu and Liu, 2004). In this case, the most
relevant features are kept and a redundancy analysis
is performed to remove redundant features.
2.2 Feature Selection Filters
We consider the three FS filters next described. The
first technique is the fast correlation-based filter
(FCBF) proposed by Yu and Liu (2003), based on
the RR framework, computing the feature-class and
feature-feature correlations. It starts by selecting a set
Union and Intersection K-Fold Feature Selection
361
of features with correlation with the class label above
some threshold (the predominant features). In the sec-
ond step, redundancy analysis finds redundant fea-
tures among the predominant ones. These redundant
features are removed, keeping the ones that are the
most relevant to the class. FCBF resorts to the sym-
metrical uncertainty (SU) (Yu and Liu, 2003) mea-
sure, defined as
SU(X
i
,X
j
) =
2I(X
i
;X
j
)
H(X
i
) + H(X
j
)
, (1)
where H(.) denotes the Shannon entropy and I(.)
denotes the mutual information (MI) (Cover and
Thomas, 2006). The SU is zero for independent ran-
dom variables and equal to one for deterministically
dependent random variables, i.e., if one is a bijective
function of the other.
The second FS technique is the Fisher ratio, a su-
pervised relevance-only method. For the i-th feature,
with C = 2, it computes the rank of the feature accord-
ing to
FiR
i
=
X
(−1)
i
− X
(1)
i
p
var(X
i
)
(−1)
+ var(X
i
)
(1)
, (2)
where X
(−1)
i
, X
(1)
i
, var(X
i
)
(−1)
, and var(X
i
)
(1)
are the
sample means and variances of feature X
i
, for the
instances of both classes. It aims to measure how
well each feature separates the two classes and is ad-
equate as a relevance metric for FS purposes. For the
multi-class case, C > 2, the FiR of feature X
i
is given
by (Duda et al., 2001; Zhao et al., 2010)
FiR
i
=
C
∑
j=1
n
(y)
j
X
( j)
i
− X
i
2
C
∑
j=1
n
(y)
j
var
X
( j)
i
, (3)
where n
(y)
j
is the number of occurrences of class j in
the n-length class label vector y, and X
( j)
i
is the sam-
ple mean of the values of X
i
whose class label is j;
finally, X
i
is the sample mean of feature X
i
.
The third FS filter is the relevance-only unsuper-
vised mean-median (MM) criterion, which ranks fea-
tures according to
MM
i
= |X
i
− median(X
i
)|. (4)
The relevance of each feature is the absolute differ-
ence between the mean and median of X
i
. This cri-
terion is based on the idea that the most relevant fea-
tures are the ones with more asymmetric distributions.
The k-fold feature selection (KFFS) filter, de-
scribed in Algorithm 1, was proposed by Ferreira and
Figueiredo (2023) and it can work with any unsuper-
vised or supervised FS filter.
KFFS follows the rationale that the importance of
a feature is proportional to the number of times it is
selected on the k-folds over the training data. It re-
quires two parameters: the number of folds k to sam-
ple the training data and the threshold T
h
to assess the
percentage of choice of a feature by the filter on the k
folds.
3 PROPOSED APPROACH
In Section 3.1, we present our key insights regard-
ing the union and intersection of feature subspaces.
The details of the proposed technique are presented
in Section 3.2.
3.1 Union and Intersection
Our proposal is built upon the idea of the union and
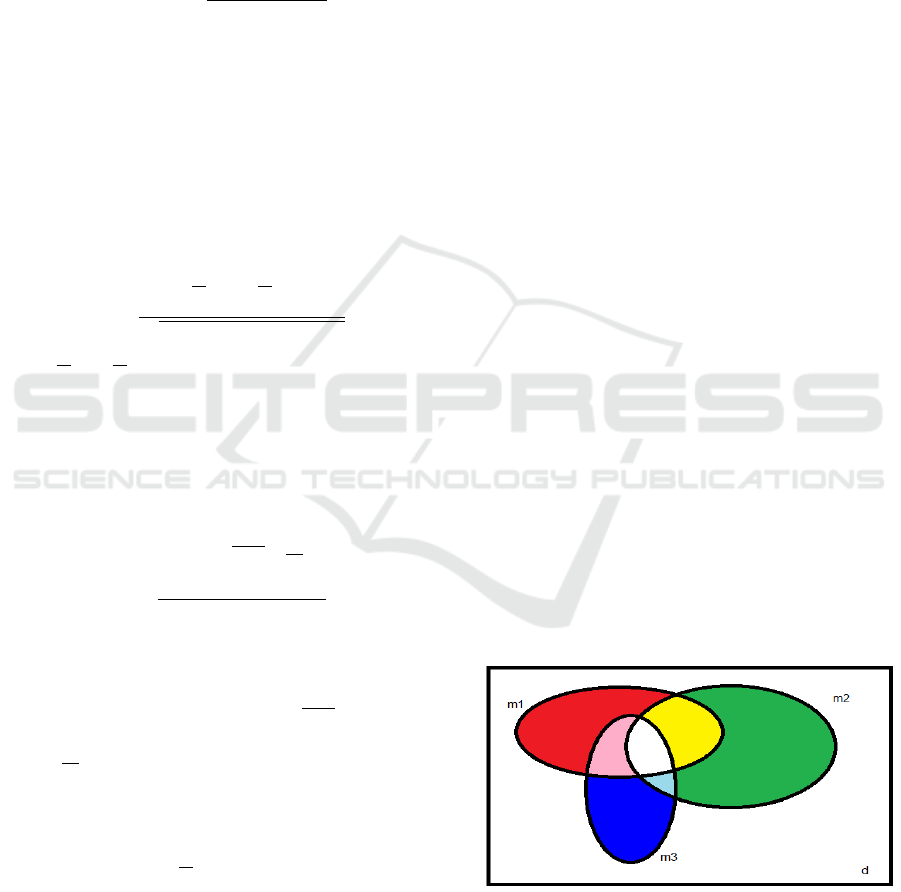
the intersection of feature subspaces, as depicted in
Figure 2. Suppose that we have a feature space with d
features and over that space we apply three different
FS filters. These filters return feature subspaces with
dimensionality m
1
, m
2
, and m
3
features, respectively.
In Figure 2, we observe the union and the intersec-
tion among these feature subspaces, using an analogy
with the additive RGB color scheme. The subspaces
selected by FS methods 1, 2, and 3 are assigned to
the primary R, G, and B colors, respectively. The
intersection of the filter subspaces is represented by
the corresponding results of the color addition on the
RGB color space. To denote the number of features in
common on the subspaces found by FS methods i and
j, we use m
i j
, with with i, j ∈ {1,2,3}; on the case of
three FS filters, we use the notation m
123
.
Figure 1: Feature subspace analysis for the case of three
FS methods, on a d-dimensional space using a visual corre-
spondence with the three primary colors.
Over these feature subspaces, we can compute
statistics to assess the relation and (dis)similarities be-
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
362

Algorithm 1: k-Fold Feature Selection (KFFS) for filter FS by Ferreira and Figueiredo (2023).
Require: X : n × d matrix, n patterns of a d-dimensional dataset.
@ f ilter : a FS filter (unsupervised or supervised).
k : an integer stating the number of folds (k ∈ {2, . ..,n}).
T
h
: a threshold (percentage) to chose the number of features.
y : n× 1 class label vector (necessary only in case of a FS supervised filter).
Ensure: idx: m−dimensional vector with the indexes of the selected features.
1: Allocate the feature counter vector (FCV ), with dimensions 1 × d, such that each position refers to a specific feature.
2: Initialize FCV
i
= 0, with i ∈ {0,... , d − 1}.
3: Compute the k data folds in the dataset (different splits into training and test data).
4: For each fold, apply @ f ilter on the training data and update FCV
i
with the number of times @ f ilter selects feature i.
5: After the k data folds are processed, convert FCV to percentage: FCV P ← FCV /k.
6: Keep the indexes of the features that have been selected at least T
h
times (expressed in percentage), idx ← FCV P ≥ T
h
.
7: Return idx (the vector with the indexes of the selected features that have been selected at least T
h
times).
tween them. The Jaccard index (JI) is one of such
metrics, being defined as
JI(A,B) =
|A ∩ B|
|A ∪ B|
, (5)
for sets A and B, where ∩ denotes intersection, ∪ de-
notes union, and |.| is the cardinality of the set. We
have 0 ≤ JI(A, B) ≤ 1. On the extreme cases, we
have: if A ∩ B =
/
0, then JI(A,B) = 0; if A ⊆ B or
B ⊆ A then JI(A, B) = 1. Other similar metrics are
the Dice-Sorenson (DS) coefficient,
DS(A,B) = 2
|A ∩ B|
|A| + |B|
, (6)
and the overlap coefficient or Szymkiewicz–Simpson
(SS) coefficient,
SS(A, B) =
|A ∩ B|
min(|A|,|B|)
, (7)
both ranging from 0 (maximally different) to 1 (max-
imally similar or one is a subset of the other).
3.2 Union and Intersection KFFS
Our proposal extends the KFFS algorithm as follows:
• Set the T
h
and k parameters of KFFS to their val-
ues; by default, we set k = 10 and T
h
= 1.
• Apply KFFS with two or three different FS fil-
ters, described in Section 2.2. We apply KFFS
(@ f ilter
1
=FCBF), KFFS (@ f ilter
2
=FiR), and
KFFS (@ f ilter
3
=MM) on the same k data folds,
using the threshold T
h
. Each filter will select dif-
ferent subsets of the input feature space, as de-
picted in Figure 2.
• Get the output indexes returned by each filter,
idx
f cb f
, idx
f ir
, and idx
mm
.
• Combine the output indexes (idx
f cb f
, idx
f ir
, and
idx
mm
) returned by the filters, with union and in-
tersection of the indexes of the selected features.
• Return the two index vectors, given by
idx
union
= idx
f cb f
∪ idx
f ir
∪ idx
mm
;
idx
intersection
= idx
f cb f
∩ idx
f ir
∩ idx
mm
.
The rationale is that by using and combining dif-
ferent filters, we are able to focus on different subsets
of the original input feature space. We also expect
that the combination of these feature subspaces will
overcome the results of each individual FS method.
The union of the feature subspaces will yield (much)
larger subspaces than the intersection of these sub-
spaces. In the intersection of the two or three sub-
spaces, we will have a small number of features which
are really relevant, since they are always selected re-
gardless the FS filter.
The unsupervised MM filter and the supervised
Fisher and FCBF FS filters were described in Sec-
tion 2.2. The MM and Fisher filters are relevance-
based methods, which select the top m most relevant
features as follows:
• Compute the MM relevance by Equation (4) or
Fisher ratio relevance by Equations (2) or (3), de-
noted as R
i
, for each feature X
i
, i ∈ {1,...,d}.
• Sort the relevance values by decreasing order.
• Compute the cumulative and normalized rele-
vance values, leading to an increasing function
whose values range to a maximum of 1.
• Keep the first top relevant m features, holding, say
90% of the accumulated relevance given by R
i
.
The FCBF filter is a relevance-redundancy based
method. We use its default parameter values.
4 EXPERIMENTAL EVALUATION
The proposed methods were evaluated with public do-
main datasets. Section 4.1 describes the datasets and
Union and Intersection K-Fold Feature Selection
363
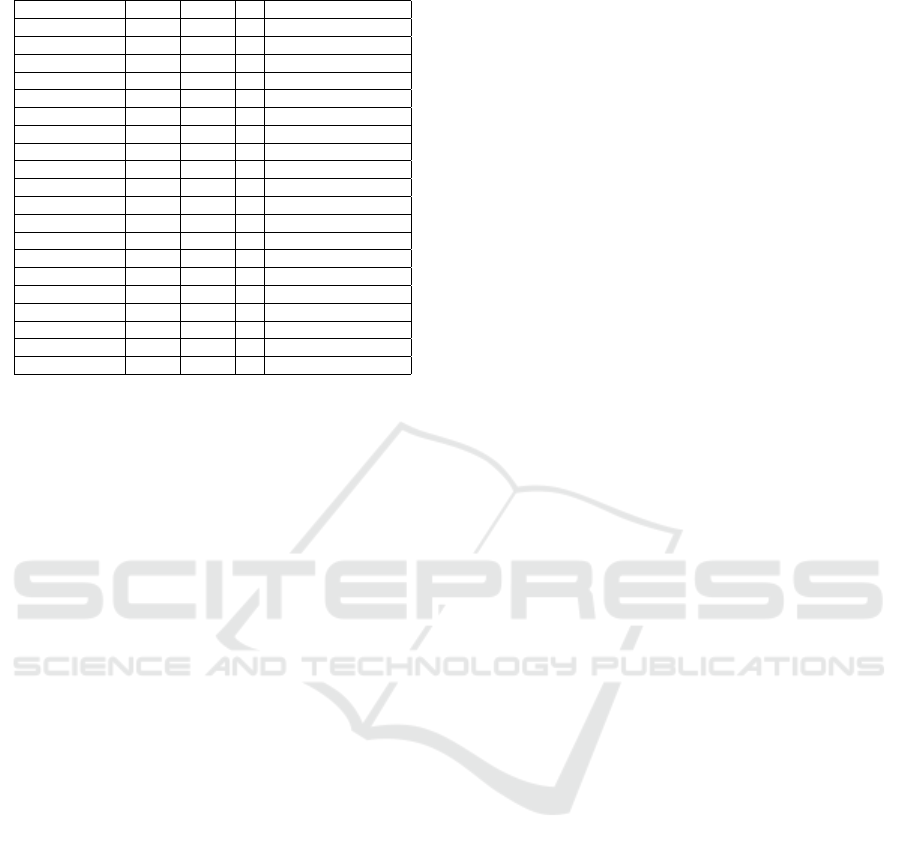
Table 1: Datasets with n instances, d features, and C classes.
Name n d C Problem/Task
Australian 690 14 2 Credit approval
Brain-Tumor-1 90 5920 5 Cancer detection
Brain-Tumor-2 50 10367 4 Cancer detection
Colon 62 2000 2 Cancer detection
Darwin 174 450 2 Alzheimer detection
Dermatology 366 34 6 Skin cancer detection
DLBCL 77 5469 2 B-cell malignancies
Drebin 15036 215 2 Malware detection
Heart 270 13 2 Heart disease
Hepatitis 155 19 2 Hepatitis survival
Ionosphere 351 34 2 Radar returns
Leukemia 72 7129 2 Leukemia detection
Leukemia-1 72 5328 3 Leukemia detection
Lymphoma 96 4026 9 Lymphoma detection
Prostate-Tumor 102 10509 2 Tumor detection
Sonar 208 60 2 Rock/Mine detection
Spambase 4601 57 2 Email spam
SRBCT 83 2308 4 Cancer detection
WDBC 569 30 2 Breast cancer
Wine 178 13 3 Wine cultivar
the evaluation metric. In Section 4.2, we report the
experimental results for the individual filters, their
union, and their intersection. In Section 4.3, we assess
the effect of changing the threshold and the number of
folds.
4.1 Datasets and Metrics
Table 1 describes the datasets used in our experi-
ments. We have gathered 20 datasets with differ-
ent types of data and problems, to assess the behav-
ior of our proposed method in different classification
task scenarios. The datasets are available from https:
//csse.szu.edu.cn/staff/zhuzx/Datasets.html, from the
Arizona State University (ASU) repository (Zhao
et al., 2010), from the UCI University of California at
Irvine (UCI) repository (Dua and Graff, 2019), https:
//archive.ics.uci.edu/ml/index.php, from the knowl-
edge extraction evolutionary learning (KEEL), https:
//sci2s.ugr.es/keel/datasets.php repository, and https:
//jundongl.github.io/scikit-feature/datasets.html.
The microarray datasets for cancer detection have
“large d, small n”, d ≫ n. Other datasets are in the
opposite situation, with n ≫ d. We have also chosen
both binary and multi-class datasets.
We use the FCBF and FiR implementation of the
ASU repository. For the MM FS filter, we have our
own implementation. We have considered the na
¨
ıve
Bayes (NB) classifier from Waikato environment for
knowledge analysis (WEKA). NB classifier is sensi-
tive to the presence of redundant features, suffering
an increase in the test-set error rate in the presence of
such features. Thus, it is useful to assess and compare
the quality of the feature subspaces found by each
method. Our key concern is to assess and compare the
adequacy of the several feature subspaces and not to
find the best classifier. For comparison purposes, we
have also used the support vector machines (SVM)
classifier.
As evaluation metric, we consider the test-set er-
ror rate, with a 10-fold cross-validation procedure.
We also analyze the size of the feature subsets.
4.2 Union and Intersection
Table 2 shows the average test set error rate (Err) and
the average number of features m, over the ten folds,
for the four combinations of unions among these sub-
spaces.
In seven out of 20 datasets, the union of filters at-
tains the best results. The best average global result
is attained by KFFS(FCBF) closely followed by the
union of the three filters. All FS filter lead to large
reduction of the dimensionality of the data.
Table 3 reports the average test set error rate (Err)
and the average number of features m, over the ten
folds, for all the possible combinations of intersec-
tions among these subspaces. The results of the indi-
vidual methods are the same as in Table 2.
For some cases, the intersection of the feature sub-
space is an empty set. In four out of the 20 datasets,
the intersection of the filters attains better results than
the use of individual filters. In generic terms, the in-
tersection of filters also yields feature subspaces of
reduced dimensionality.
4.3 Parameter Sensitivity
We analyze the effect of changing the threshold T
h
in
KFFS(FCBF), KFFS(FiR) and their union and inter-
section for the Prostate-Tumor dataset, in Figure 2.
The FS approaches improve significantly the results
of the baseline approach, with a consistent behavior.
In KFFS, as we increase the threshold the dimension-
ality of the selected feature space decreases.
Table 4 reports the best threshold value for each
dataset. We have made a grid search over all the pos-
sible threshold values from 0 to 100, and for each of
the four filters KFFS(FCBF), KFFS(Fisher), and their
union and intersection, we have recorded the highest
threshold (fewer features) with the lowest test set er-
ror rate by the SVM classifier.
We now analyze the effect of changing the num-
ber of folds k in KFFS, for a fixed threshold T
h
.
The goal is to assess the sensitivity of our proposed
method with the number of sampling folds on the
training data. In Figure 3, we assess the test set er-
ror rate of the SVM classifier with 10-fold CV, on
the DLBCL dataset, with ten different values of k ∈
{n/10,2n/10, .. ., n} and a fixed T
h
= 50.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
364
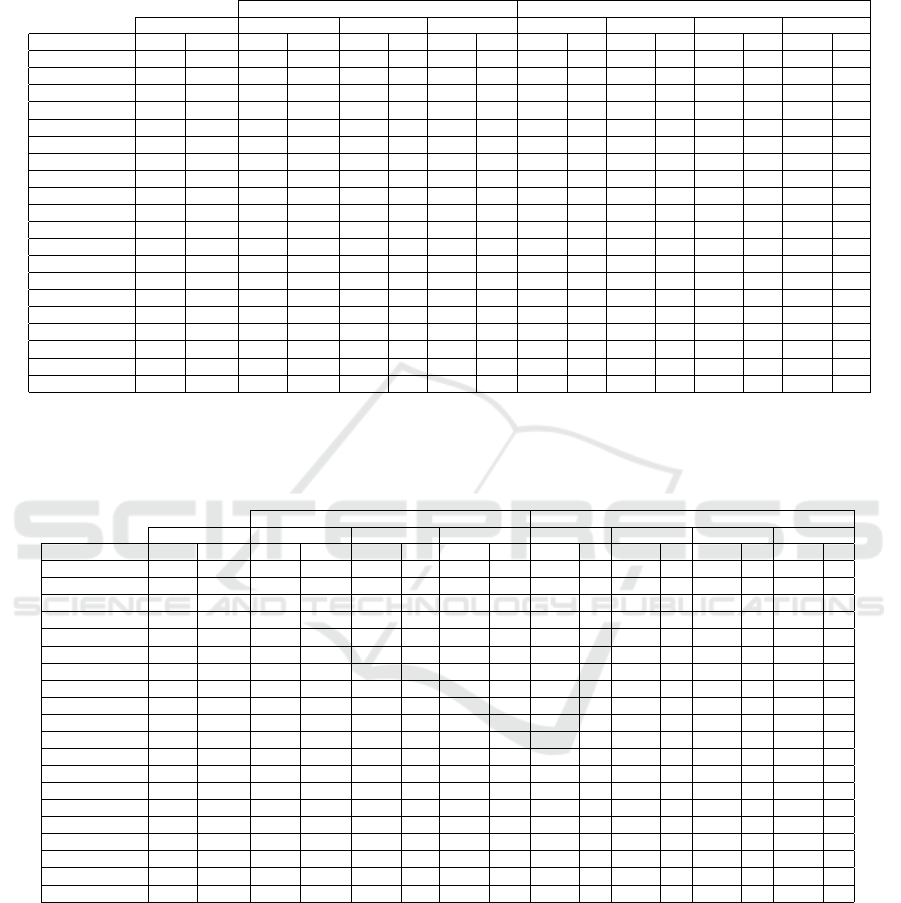
Table 2: Union evaluation. The average values of test set error rate (Err, in %) and the average number of features m for each
individual FS filter and their union, on the ten folds of 10-fold CV, for all the benchmark datasets. We use KFFS with k = 10
and T
h
= 1 with @ f ilter
1
= FCBF, @ f ilter
2
= FiR, and @ f ilter
3
= MM. The lower Err is in boldface. Regarding the error
rates, the Friedman test p-value is p = 0.0010340240 (≤ 0.05), thus having statistical significance.
Individual Filters Union of Filters
Baseline NB KFFS(FCBF) KFFS(FiR) KFFS(MM) ∪
12
∪
13
∪
23
∪
123
Dataset Err d Err m Err m Err m Err m Err m Err m Err m
Australian 23.48 14 24.06 8 23.48 9 24.93 9 23.33 10 23.91 11 23.62 12 23.62 12
Brain-Tumor-1 10.00 5920 12.22 473 18.89 205 43.33 1 12.22 617 12.22 474 18.89 206 12.22 618
Brain-Tumor-2 32.00 10367 28.00 359 24.00 205 34.00 121 22.00 535 24.00 474 32.00 303 20.00 630
Colon 40.48 2000 19.05 55 17.62 86 52.14 1 21.19 114 19.05 57 15.95 88 17.86 116
Darwin 12.68 450 12.06 112 13.20 57 14.38 62 11.50 139 14.41 151 12.65 104 11.50 174
Dermatology 2.80 34 3.63 19 26.53 8 2.80 29 3.63 23 2.80 31 2.80 30 2.80 31
DLBCL 18.21 5469 6.43 225 9.11 116 15.36 2 10.54 292 6.43 226 10.54 117 10.54 292
Drebin 16.64 215 8.73 17 19.89 28 21.56 48 18.24 42 20.12 62 20.69 52 19.14 66
Heart 15.56 13 16.67 8 15.19 10 17.78 10 15.56 11 15.19 11 15.19 12 15.19 12
Hepatitis 15.42 19 17.42 9 17.33 12 19.33 17 16.08 12 16.75 18 18.00 18 16.08 18
Ionosphere 18.81 34 8.83 15 18.25 16 19.38 23 16.25 23 17.10 27 19.10 23 16.81 27
Leukemia 1.43 7129 2.86 171 4.29 142 33.39 2 2.86 256 2.86 173 4.29 144 2.86 258
Leukemia-1 4.29 5327 5.71 204 4.29 149 54.46 2 4.29 301 5.71 207 4.29 152 4.29 304
Lymphoma 24.00 4026 23.11 848 18.00 128 15.56 112 19.89 904 22.00 921 12.67 218 20.89 964
Prostate-Tumor 37.09 10509 9.64 257 8.73 114 32.09 100 10.55 320 14.55 354 11.55 211 12.55 415
Sonar 32.74 60 34.67 18 34.67 20 31.64 14 35.62 25 30.81 31 33.21 32 32.74 36
Spambase 20.73 54 23.63 18 13.45 14 21.45 28 21.02 24 20.28 35 20.89 33 20.28 37
SRBCT 1.11 2308 0.00 203 1.11 145 6.11 118 1.11 267 0.00 297 1.11 239 1.11 351
WDBC 6.67 30 4.92 11 6.85 14 7.20 13 6.14 20 5.79 18 6.49 18 6.14 22
Wine 2.78 13 2.22 10 5.56 5 3.33 12 2.22 10 2.78 13 3.33 12 2.78 13
Table 3: Intersection evaluation. The average values of test set error rate (Err, in %) and the average number of features m
for each individual FS filter and their intersection, on the ten folds of 10-fold CV, for all the benchmark datasets. We use
KFFS with k = 10 and T
h
= 1 with @ f ilter
1
= FCBF, @ f ilter
2
= FiR, and @ f ilter
3
= MM. The lower Err is in boldface.
Regarding the error rates, the Friedman test p-value is p = 0.0010340240 (≤ 0.05), thus having statistical significance.
Individual Filters Intersection of Filters
Baseline NB KFFS(FCBF) KFFS(FiR) KFFS(MM) ∩
12
∩
13
∩
23
∩
123
Dataset Err d Err m Err m Err m Err m Err m Err m Err m
Australian 23.48 14 24.06 8 23.48 9 24.93 9 24.06 7 25.65 6 24.78 6 25.80 5
Brain-Tumor-1 10.00 5920 12.22 473 18.89 205 43.33 1 20.00 62 – 0 – 0 – 0
Brain-Tumor-2 32.00 10367 28.00 359 24.00 205 34.00 121 30.00 30 34.00 6 38.00 23 46.00 3
Colon 40.48 2000 19.05 55 17.62 86 52.14 1 19.05 27 – 0 – 0 – 0
Darwin 12.68 450 12.06 112 13.20 57 14.38 62 12.58 30 13.73 23 13.17 15 16.50 11
Dermatology 2.80 34 3.63 19 26.53 8 2.80 29 27.09 5 5.03 17 30.16 7 30.71 4
DLBCL 18.21 5469 6.43 225 9.11 116 15.36 2 5.00 50 19.64 1 – 0 – 0
Drebin 16.64 215 8.73 17 19.89 28 21.56 48 11.34 3 11.79 2 21.14 24 11.79 2
Heart 15.56 13 16.67 8 15.19 10 17.78 10 16.67 8 17.41 7 18.52 9 18.15 7
Hepatitis 15.42 19 17.42 9 17.33 12 19.33 17 19.92 8 17.38 8 17.96 11 19.88 7
Ionosphere 18.81 34 8.83 15 18.25 16 19.38 23 12.83 8 11.12 10 19.10 15 13.69 7
Leukemia 1.43 7129 2.86 171 4.29 142 33.39 2 2.86 57 – 0 – 0 – 0
Leukemia-1 4.29 5327 5.71 204 4.29 149 54.46 2 4.11 52 – 0 – 0 – 0
Lymphoma 24.00 4026 23.11 848 18.00 128 15.56 112 19.11 72 13.67 39 16.67 22 20.89 9
Prostate-Tumor 37.09 10509 9.64 257 8.73 114 32.09 100 7.73 51 16.73 3 19.64 2 13.82 1
Sonar 32.74 60 34.67 18 34.67 20 31.64 14 35.60 14 40.90 2 40.86 2 41.86 1
Spambase 20.73 54 23.63 18 13.45 14 21.45 28 15.89 8 25.06 11 14.63 10 16.91 6
SRBCT 1.11 2308 0.00 203 1.11 145 6.11 118 1.25 81 4.86 24 3.61 24 7.22 14
WDBC 6.67 30 4.92 11 6.85 14 7.20 13 5.79 5 6.85 5 8.78 9 8.43 4
Wine 2.78 13 2.22 10 5.56 5 3.33 12 5.56 5 2.78 10 5.56 5 5.56 5
The number of folds k has a large impact on the
end result for all filters. For lower values of k, we have
a non-stationary behavior of the error rate curve. Af-
ter a sufficiently large value of k, we observe a more
stable behavior on the error rate. These results show
that, for a specific dataset and problem, one should
fine-tune both the T
h
and k parameters to have better
results.
5 CONCLUSIONS
In this paper, we have extended the KFFS filter algo-
rithm by performing union and intersection of the in-
dividual feature subspaces of two and three heteroge-
neous FS filters. We have considered two supervised
FS filters (FCBF and FiR) and one unsupervised filter
(MM). Two of these filters are relevance based (FiR
Union and Intersection K-Fold Feature Selection
365

Table 4: The best test set error rate (Err, in %), the corresponding average number of features m and Threshold, T
h
, for
KFFS(FCBF), KFFS(Fisher), and their union and intersection, for all the benchmark datasets. We use KFFS with k = 10 and
the SVM classifier. The best result is in boldface.
Individual Filters Union Intersection
Baseline SVM KFFS(FCBF) KFFS(FiR) ∪
12
∩
12
Dataset Err d Err m T
h
Err m T
h
Err m T
h
Err m T
h
Australian 14.49 14 14.49 4 91 14.49 7 81 14.49 8 81 14.49 3 91
Brain-Tumor-1 10.00 5920 10.00 75 31 10.00 5920 0 10.00 158 31 10.00 5920 0
Brain-Tumor-2 20.00 10367 20.00 10367 0 18.00 92 21 16.00 219 11 18.00 30 1
Colon 13.10 2000 11.43 17 21 13.10 17 91 11.43 117 1 13.10 2000 0
Darwin 17.12 450 16.60 35 31 14.38 37 11 16.57 70 21 17.12 450 0
Dermatology 3.36 34 3.08 20 1 3.36 34 0 2.79 17 51 3.36 34 0
DLBCL 2.50 5469 2.50 42 41 2.50 5469 0 2.50 79 41 2.50 25 11
Drebin 2.23 215 2.23 215 0 2.23 215 0 2.23 215 0 2.23 215 0
Heart 15.93 13 14.07 8 1 14.07 10 1 14.07 10 1 14.07 8 1
Hepatitis 23.29 19 19.38 5 61 18.17 10 21 17.46 10 61 18.79 7 11
Ionosphere 11.42 34 11.42 34 0 11.42 34 0 11.13 22 1 11.42 34 0
Leukemia 1.43 7129 1.43 7129 0 1.43 7129 0 1.43 7129 0 1.43 7129 0
Leukemia-1 1.43 5327 1.43 5327 0 1.43 5327 0 1.43 5327 0 1.43 5327 0
Lymphoma 4.33 4026 4.33 80 61 4.33 4026 0 4.33 93 71 4.33 4026 0
Prostate-Tumor 8.00 10509 6.00 24 61 6.00 54 51 5.00 65 61 4.00 48 1
Sonar 21.71 60 21.19 9 41 21.24 18 11 21.24 21 11 21.71 60 0
Spambase 10.06 54 10.06 54 0 10.06 54 0 10.06 54 0 10.06 54 0
SRBCT 0.00 2308 0.00 54 41 0.00 56 91 0.00 61 91 0.00 31 31
WDBC 2.28 30 2.28 30 0 2.28 30 0 1.93 17 21 2.28 30 0
Wine 0.56 13 0.56 8 91 0.56 13 0 0.56 9 81 0.56 13 0
Figure 2: Test set error rate of the NB classifier with
10-fold CV, as a function of the threshold in KFFS, for
KFFS(FCBF), KFFS(Fisher), and their Union and Intersec-
tion, with k = 10 on the Prostate-Tumor dataset.
and MM) while FCBF follows the RR framework.
Our experiments on 20 datasets with diverse types
of data and problems show that the union of the fea-
ture subsets typically attains better results than the
individual filters. The intersection also attains ade-
quate results, yielding human manageable subsets of
features allowing for explainability and interpretabil-
ity. By setting properly the threshold of the KFFS
algorithm, we can control the dimensionality of the
feature subspaces, reduced in such a way that allows
for the domain expert (e.g., a medical doctor) to fo-
cus on the interpretation of the resulting variables.
Figure 3: Test set error rate of the SVM classifier with 10-
fold CV, as a function of the number of folds in KFFS, for
KFFS(FCBF), KFFS(Fisher), and their Union and Intersec-
tion, with T
h
= 50 on the DLBCL dataset.
However, in some cases, the subspace intersection is
empty. The dimensionality of the subspace resulting
from the intersection is typically much lower, as com-
pared to the one from the union. When dealing with
high-dimensional data, it is often the case that FS fil-
ters select different regions of the feature subspace.
As future work, we aim to fine-tune the pa-
rameters of the method for each dataset or type of
data/problem, individually. We will also explore the
use of different thresholds per filter.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
366

ACKNOWLEDGEMENTS
This research was supported by Instituto
Polit
´
ecnico de Lisboa (IPL) under Grant
IPL/IDI&CA2024/ML4EP ISEL.
REFERENCES
Alipoor, G., Mirbagheri, S., Moosavi, S., and Cruz, S.
(2022). Incipient detection of stator inter-turn short-
circuit faults in a doubly-fed induction generator using
deep learning. IET Electric Power Applications.
Arya, L. and Gupta, G. P. (2023). Ensemble filter-based
feature selection model for cyber attack detection in
industrial internet of things. In 2023 9th International
Conference on Advanced Computing and Communi-
cation Systems (ICACCS), volume 1, pages 834–840.
Bashir, S., Khattak, I. U., Khan, A., Khan, F. H., Gani,
A., and Shiraz, M. (2022). A novel feature selec-
tion method for classification of medical data using
filters, wrappers, and embedded approaches. Com-
plexity, 2022(1):1–12.
Bol
´
on-Canedo, V. and Alonso-Betanzos, A. (2019). Ensem-
bles for feature selection: A review and future trends.
Information Fusion, 52:1–12.
Bolon-Canedo, V., Sanchez-Marono, N., and Alonso-
Betanzos, A. (2015). Feature Selection for High-
Dimensional Data. Springer.
Chamlal, H., Ouaderhman, T., and Rebbah, F. (2022). A hy-
brid feature selection approach for microarray datasets
using graph theoretic-based method. Information Sci-
ences, 615:449–474.
Cover, T. and Thomas, J. (2006). Elements of information
theory. John Wiley & Sons, second edition.
Damtew, Y. G., Chen, H., and Yuan, Z. (2023). Hetero-
geneous ensemble feature selection for network intru-
sion detection system. Int. J. Comput. Intell. Syst.,
16(1).
Dhal, P. and Azad, C. (2022). A comprehensive survey on
feature selection in the various fields of machine learn-
ing. Applied Intelligence, 52(4):4543–45810.
Dua, D. and Graff, C. (2019). UCI machine learning repos-
itory.
Duda, R., Hart, P., and Stork, D. (2001). Pattern classifica-
tion. John Wiley & Sons, second edition.
Escolano, F., Suau, P., and Bonev, B. (2009). Information
Theory in Computer Vision and Pattern Recognition.
Springer.
Ferreira, A. and Figueiredo, M. (2023). Leveraging explain-
ability with k-fold feature selection. In 12th Inter-
national Conference on Pattern Recognition Applica-
tions and Methods (ICPRAM), pages 458–465.
Guyon, I. and Elisseeff, A. (2003). An introduction to vari-
able and feature selection. Journal of Machine Learn-
ing Research (JMLR), 3:1157–1182.
Guyon, I., Gunn, S., Nikravesh, M., and Zadeh (Editors), L.
(2006). Feature extraction, foundations and applica-
tions. Springer.
Hastie, T., Tibshirani, R., and Friedman, J. (2009). The El-
ements of Statistical Learning. Springer, 2nd edition.
Huynh-Cam, T.-T., Nalluri, V., Chen, L.-S., and Yang, Y.-
Y. (2022). IS-DT: A new feature selection method for
determining the important features in programmatic
buying. Big Data and Cognitive Computing, 6(4).
Jeon, Y. and Hwang, G. (2023). Feature selection with
scalable variational gaussian process via sensitivity
analysis based on L2 divergence. Neurocomputing,
518:577–592.
Mochammad, S., Noh, Y., Kang, Y.-J., Park, S., Lee, J.,
and Chin, S. (2022). Multi-filter clustering fusion for
feature selection in rotating machinery fault classifi-
cation. Sensors, 22(6).
Pudjihartono, N., Fadason, T., Kempa-Liehr, A., and
O’Sullivan, J. (2022a). A review of feature selection
methods for machine learning-based disease risk pre-
diction. Frontiers in Bioinformatics, 2:927312.
Pudjihartono, N., Fadason, T., Kempa-Liehr, A. W., and
O’Sullivan, J. M. (2022b). A review of feature selec-
tion methods for machine learning-based disease risk
prediction. Front. Bioinform., 2:927312.
Remeseiro, B. and Bolon-Canedo, V. (2019). A review
of feature selection methods in medical applications.
Computers in Biology and Medicine, 112:103375.
Seijo-Pardo, B., Porto-D
´
ıaz, I., Bol
´
on-Canedo, V., and
Alonso-Betanzos, A. (2017). Ensemble feature selec-
tion: Homogeneous and heterogeneous approaches.
Knowledge-Based Systems, 118:124–139.
Xu, Y., Liu, Y., and Ma, J. (2022). Detection and de-
fense against DDoS attack on SDN controller based
on feature selection. In Chen, X., Huang, X., and
Kutyłowski, M., editors, Security and Privacy in So-
cial Networks and Big Data, pages 247–263, Singa-
pore. Springer Nature Singapore.
Yu, L. and Liu, H. (2003). Feature selection for high-
dimensional data: a fast correlation-based filter solu-
tion. In Proceedings of the International Conference
on Machine Learning (ICML), pages 856–863.
Yu, L. and Liu, H. (2004). Efficient feature selection via
analysis of relevance and redundancy. Journal of Ma-
chine Learning Research (JMLR), 5:1205–1224.
Zhao, Z., Morstatter, F., Sharma, S., Alelyani, S., Anand,
A., and Liu, H. (2010). Advancing feature selection
research - ASU feature selection repository. Techni-
cal report, Computer Science & Engineering, Arizona
State University.
´
Alvarez Est
´
evez, D., S
´
anchez-Maro
˜
no, N., Alonso-
Betanzos, A., and Moret-Bonillo, V. (2011). Reducing
dimensionality in a database of sleep EEG arousals.
Expert Systems with Applications, 38(6):7746–7754.
Union and Intersection K-Fold Feature Selection
367
