
Classification of Oral Cancer and Leukoplakia Using Oral Images and
Deep Learning with Multi-Scale Random Crop Self-Training
Itsuki Hamada
1
, Takaaki Ohkawauchi
2
, Chisa Shibayama
3
, Kitaro Yoshimitsu
4
, Nobuyuki Kaibuchi
3
,
Katsuhisa Sakaguchi
5
, Toshihiro Okamoto
3
and Jun Ohya
1
1
Department of Modern Mechanical Engineering, Waseda University, Tokyo, Japan
2
College of Humanities and Sciences, Nihon University, Tokyo, Japan
3
Department of Oral and Maxillofacial Surgery, Tokyo Women’s Medical University, Tokyo, Japan
4
Institute of Advanced Biomedical Engineering and Science, Faculty of Advanced Techno-Surgery,
Tokyo Women’s Medical University, Tokyo, Japan
5
Department of Medical Engineering, Tokyo City University, Tokyo, Japan
Keywords:
Medical Image Analysis, Diagnostic Imaging, Oral Cancer, Leukoplakia, Multi-Scale Random Crop
Self-Training, Semi-Supervised Learning, Vision Transformer, MixUp, Dermoscopy Imaging.
Abstract:
This paper proposes Multi-Scale Random Crop Self-Training (MSRCST) for classifying oral cancers and
leukoplakia using oral images acquired by our dermoscope. MSRCST comprises the following three key
modules: (1) Multi-Scale Random Crop, which extracts image patches at various scales from high-resolution
images, preserving both local details and global contextual information essential for accurate classification, (2)
Selection based on Confidence, which employs a teacher model to assign confidence scores to each cropped
patch, selecting only those with high confidence for further training and ensuring the model focusing on di-
agnostically relevant features, (3) Iteration of Self-training, which iteratively retrains the model using the
selected high-confidence, pseudo-labeled data, progressively enhancing accuracy. In our experiments, we ap-
plied MSRCST to classify images of oral cancer and leukoplakia. When combined with MixUp data augmen-
tation, MSRCST achieved an average classification accuracy of 71.71%, outperforming traditional resizing
and random cropping methods. Additionally, it effectively reduced misclassification rates, as demonstrated by
improved confusion matrices, thereby enhancing diagnostic reliability.
1 INTRODUCTION
In Japan, 7,827 deaths from oral and pharyngeal can-
cer were reported in 2020 (National Cancer Center
Japan). Early detection is critical, as it allows for less
invasive treatments and better outcomes, while late-
stage cancer significantly lowers survival rates and in-
creases complications (Japan Society for Oral Cancer
Elimination). However, early symptoms often resem-
ble those of leukoplakia, making diagnosis by visual
and tactile examinations challenging, especially for
non-specialists. Existing methods like iodine stain-
ing and magnifying endoscopy (Nomura et al., 2008;
Shibahara et al., 2014) are limited by cost and patient
discomfort.
Deep learning technologies, such as CNNs
(Krizhevsky et al., 2012), have shown promise in
medical imaging, enabling high-accuracy classifica-
Table 1: Summary of Cases and Image Data. Overview
of the number of cases and collected images for oral cancer
and leukoplakia.
Class Number of Cases Number of Images
Oral Cancer 15 567
Leukoplakia 13 391
tion and anomaly detection (Rajpurkar et al., 2017;
Litjens et al., 2017). In this study, we created a dataset
by extracting images with a resolution of 640×480
from videos of 28 cases captured using an oral dermo-
scope developed by Tokyo Women’s Medical Univer-
sity. Using this dataset, we applied deep learning to
diagnose oral cancer. We fine-tuned pre-trained mod-
els, such as ResNet (He et al., 2015) and ViT (Doso-
vitskiy et al., 2021), and evaluated the effectiveness
of MixUp (Zhang et al., 2018) in improving classifi-
780
Hamada, I., Ohkawauchi, T., Shibayama, C., Yoshimitsu, K., Kaibuchi, N., Sakaguchi, K., Okamoto, T. and Ohya, J.
Classification of Oral Cancer and Leukoplakia Using Oral Images and Deep Learning with Multi-Scale Random Crop Self-Training.
DOI: 10.5220/0013296500003905
In Proceedings of the 14th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2025), pages 780-787
ISBN: 978-989-758-730-6; ISSN: 2184-4313
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

Figure 1: Representative Images of Oral Cancer and
Leukoplakia. An example of a 640×480 pixel image is
shown, depicting lesions of oral cancer and leukoplakia.
The three images on the left display oral cancer, while those
on the right show leukoplakia.
Figure 2: Comparison of Resizing and Cropping High-
Resolution Images. Illustration of the differences between
resizing and cropping medical images, showing the poten-
tial impact on information loss and diagnostic accuracy.
cation accuracy. Table 1 provides an overview, and
Figure 1 shows sample images.
In deep learning, the quantity and quality of train-
ing data significantly affect a model’s generalization.
Large, high-quality datasets improve accuracy and
prevent overfitting, while limited or mislabeled data
can degrade performance. Fine-tuning pre-trained
models, like those trained on ImageNet, enhances
generalization, reduces overfitting, and compensates
for data limitations. However, resizing or cropping
high-resolution images (e.g., 640×480 to 224×224
pixels) to meet input constraints can, as shown in Fig-
ure 2, result in the loss of critical details or context
needed to distinguish cancer from leukoplakia.
To address these challenges, we propose Multi-
Scale Random Crop Self-Training (MSRCST), a
semi-supervised method. It generates multiple
cropped images at varying scales from a single high-
resolution image, ranks them using confidence scores
from a teacher model, and assigns pseudo-labels to
the top images. This process iteratively creates a
diverse dataset, capturing diagnostic-critical regions
while preserving important features for training.
2 TECHNOLOGIES USED IN
THIS PAPER
ViT (Vision Transformer) (Dosovitskiy et al., 2021)
is a novel architecture that leverages self-attention
mechanisms (Vaswani et al., 2017) to process images
by dividing them into patches. By fine-tuning pre-
trained models, it enables efficient learning and im-
proves accuracy.
MixUp (Zhang et al., 2018) is a data augmenta-
tion technique that generates new samples by linearly
combining images and their labels. This approach
broadens the data distribution, prevents overfitting,
and enhances the model’s robustness. In this study,
we aim to build a more generalized model by com-
bining MixUp with standard data augmentation tech-
niques.
Semi-supervised Learning (Lee, 2013; Xie et al.,
2020) combines a small amount of labeled data with
a large amount of unlabeled data for training. Among
these methods, Self-training involves training an ini-
tial model with labeled data, assigning pseudo-labels
to unlabeled data, and retraining iteratively. This ap-
proach reduces labeling costs while enhancing the
model’s generalization performance.
3 APPROACH
This study proposes a method called Multi-Scale Ran-
dom Crop Self-Training (MSRCST) to effectively uti-
lize high-resolution medical images for classifying
oral cancer and leukoplakia. MSRCST consists of
two phases: “Teacher Model Training” and “Itera-
tive Self-Training” (indicated in blue in Figure 3). To
address the limitations of conventional resizing and
cropping methods, MSRCST incorporates two key
modules: Multi-Scale Random Crop and Selection
Based on Confidence (highlighted in red in Figure 3).
Additionally, a Confidence-Based Evaluation method
is introduced to accurately assess the performance of
the trained models.
Classification of Oral Cancer and Leukoplakia Using Oral Images and Deep Learning with Multi-Scale Random Crop Self-Training
781
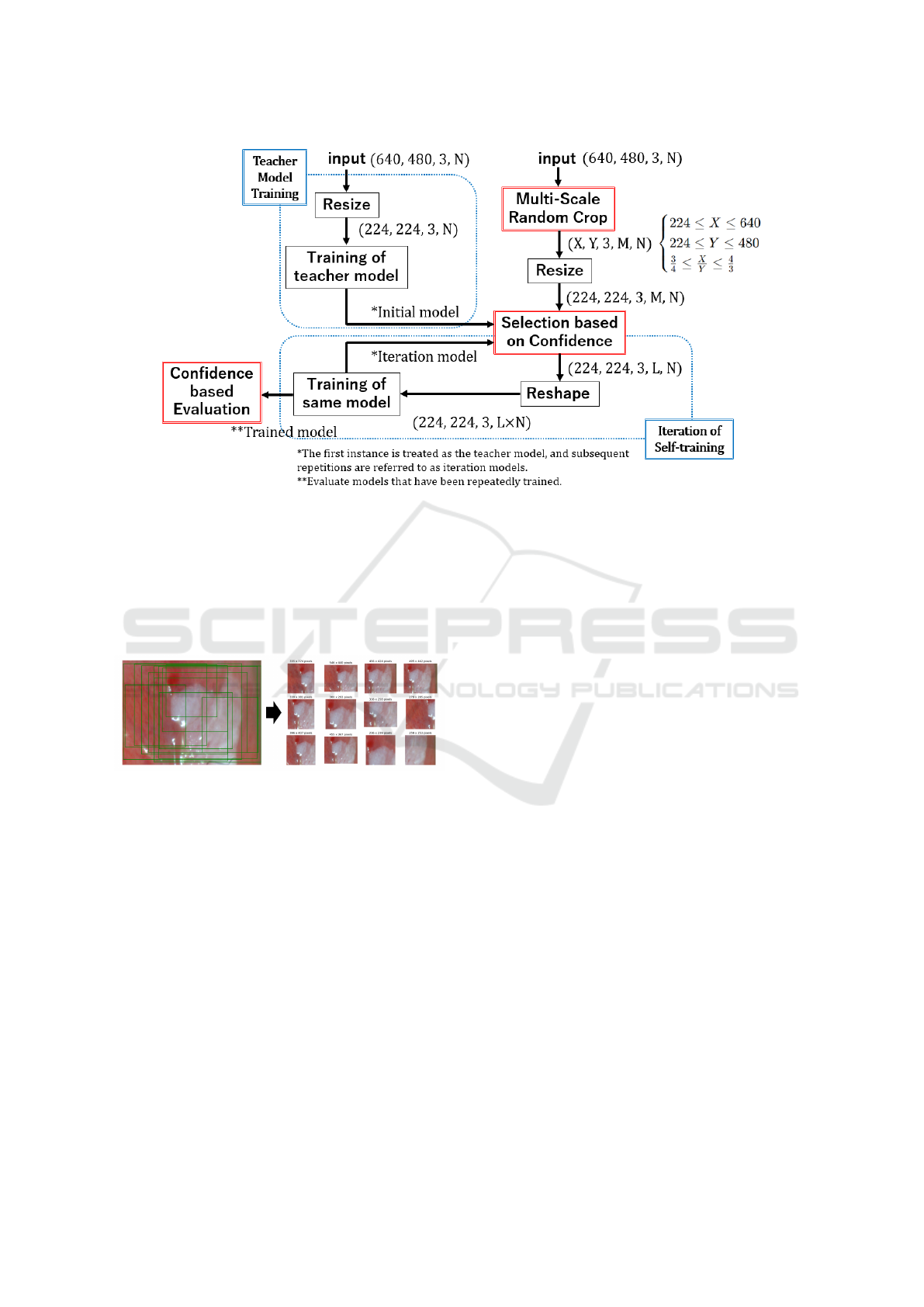
Figure 3: MSRCST Framework Overview. This figure illustrates the workflow of Multi-Scale Random Crop Self-
Training (MSRCST), which includes tensor transformations and iterative processes. Input image tensors are defined as
(horizontal pixels, vertical pixels, channels, training samples). The process starts with Teacher Model Training, where input
images (640, 480, 3, N) are resized to (224, 224, 3, N) for training to establish the teacher model’s foundational performance.
Next, Multi-Scale Random Crop generates patches (X, Y, 3, M, N), where X and Y represent the horizontal and vertical
lengths of the cropped images, respectively, and M is the number of patches per image. These patches are resized and filtered
by confidence to (224, 224, 3, L, N), with L as the number of selected high-confidence patches. The tensor is reshaped into
(224, 224, 3, L × N) for training.During iterative self-training, only high-confidence patches are used to refine the model. This
process is repeated to enhance performance, concluding with a Confidence-Based Evaluation step.
Figure 4: Multi-Scale Random Crop (MSRC) Technique.
Illustration of the MSRC method applied to high-resolution
images, demonstrating how diverse patches are extracted
from various scales across the entire image to enhance
model training.
3.1 Challenges with Resizing and
Cropping
High-resolution medical images (640×480 pixels)
contain critical diagnostic information in both global
context and fine-grained local details. However, com-
monly used pre-trained models such as ResNet and
Vision Transformer (ViT) require input images to be
resized to 224×224 pixels, which introduces the fol-
lowing challenges:
• Resizing: While resizing ensures that all regions
are included, it reduces image resolution, causing
the loss of fine-grained features critical for distin-
guishing oral cancer from leukoplakia.
• Cropping: Cropping preserves high-resolution
details and enhances dataset diversity. However,
random cropping risks omitting essential diagnos-
tic regions, leading to the loss of contextual infor-
mation.
To overcome these challenges, MSRCST combines
the strengths of resizing and cropping while minimiz-
ing their drawbacks.
3.2 Workflow of MSRCST
MSRCST consists of the following two phases:
Teacher Model Training. In the first phase, a
teacher model is trained using resized images
(224×224 pixels) that include all diagnostic re-
gions. While resizing reduces resolution, it allows
the teacher model to capture overall patterns, which
serves as a foundation for confidence score calcula-
tion in the next phase.
Iterative Self-Training. In this phase, Multi-Scale
Random Crop generates high-resolution patches at
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
782
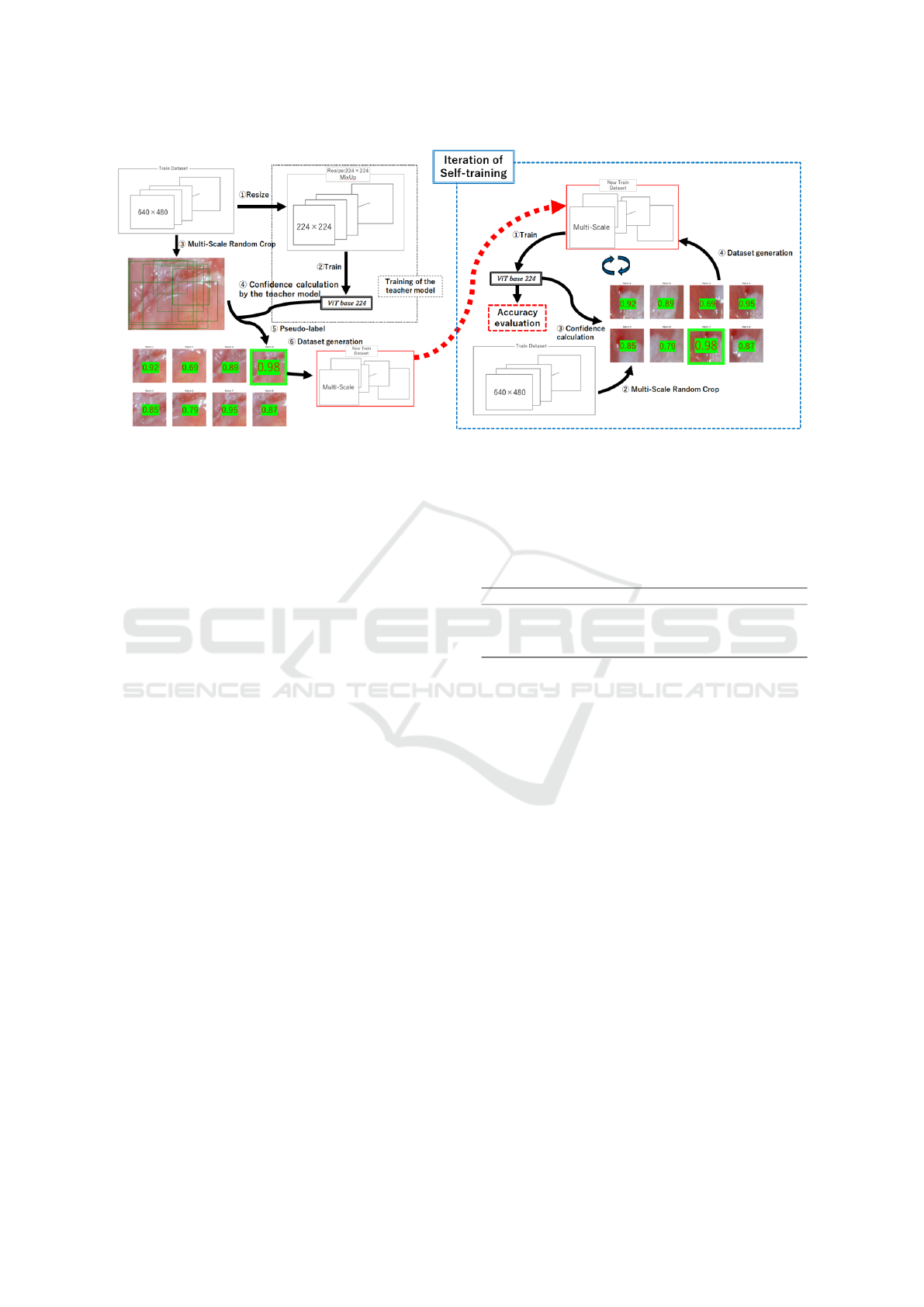
Figure 5: Overview of Multi-Scale Random Crop Self-Training (MSRCST). This figure illustrates the key components of
MSRCST, including Selection Based on Confidence and the Iterative Process of Self-Training. The diagram demonstrates
how high-confidence pseudo-labeled data are selected and used in an iterative cycle to progressively enhance the model’s
classification accuracy.
varying scales. The teacher model calculates con-
fidence scores for these patches, and only high-
confidence patches are selected for retraining. This
iterative process progressively improves the model’s
performance.
3.3 Key Components
• Multi-Scale Random Crop (MSRC): A data
augmentation technique that generates patches
at multiple scales from high-resolution images.
Each patch has a minimum side length of 224
pixels and maintains an aspect ratio between 3:4
and 4:3 to preserve contextual relationships. This
method retains fine-grained features lost during
resizing and introduces diverse perspectives into
training (Figure 4).
• Selection Based on Confidence: Confidence
scores assigned by the teacher model are used
to select patches likely to contain critical diag-
nostic regions. This process eliminates irrelevant
patches, improving the quality of training data and
reducing the risk of mislearning.
• Iterative Self-Training: Selected high-
confidence patches are used to retrain the
model, and new patches are generated for subse-
quent iterations. This iterative process enables
the model to progressively learn diagnostically
important features with greater precision.
Table 2: Data Summary of Oral Cancer and Leuko-
plakia Cases. This shows the number of cases of oral can-
cer and leukoplakia in each group, along with the corre-
sponding data count, used during cross-validation.
Group Oral Cancer Cases Leukoplakia Cases Oral Cancer Data Leukoplakia Data
1 3 4 132 109
2 4 3 154 85
3 3 4 109 132
4 5 2 172 65
3.4 Confidence-Based Evaluation
To accurately evaluate the model’s performance, a
Confidence-Based Evaluation method is introduced.
This method applies Multi-Scale Random Crop to
test images, generating multiple patches. The model
predicts confidence scores for these patches, and the
patch with the highest score determines the final pre-
diction for the entire test image. This ensures fair
evaluation by aligning with the model’s training fo-
cus.
3.5 Advantages of MSRCST
The main advantage of MSRCST lies in its ability to
generate multi-scale data from high-resolution images
and select diagnostically important patches based on
confidence. This method minimizes the risk of mis-
learning while effectively balancing the learning of
local and global features. Additionally, through itera-
tive self-training, it enhances the model’s generaliza-
tion performance and diagnostic accuracy. Further-
more, MSRCST outperforms conventional methods,
leveraging high-resolution medical images to enable
Classification of Oral Cancer and Leukoplakia Using Oral Images and Deep Learning with Multi-Scale Random Crop Self-Training
783

precise and accurate diagnosis.
4 EXPERIMENTS
4.1 Dataset
As mentioned in Chapter 1, this dataset was created
using images extracted from videos captured with
an oral dermoscope developed by Tokyo Women’s
Medical University. The images were resized to
640×480 pixels and labeled by specialists as either
”oral cancer” or ”leukoplakia.” The dataset includes
15 cases of oral cancer (567 images) and 13 cases of
leukoplakia (391 images) (Table 1). To enhance the
model’s generalization performance, data augmenta-
tion techniques such as random rotation and flipping
were applied.
4.2 Cross-Validation Method
Group K-Fold cross-validation was used to validate
the proposed method, ensuring that data from the
same case did not appear in both training and vali-
dation sets. The dataset was divided into four groups,
maintaining balanced distributions of oral cancer and
leukoplakia. Each group was used once for valida-
tion, while the remaining groups were used for train-
ing. Accuracy was weighted and averaged across
folds (Table 2), ensuring the model was not biased
towards specific data.
4.3 ResNet and ViT: Effect of MixUp
Before testing MSRCST, ResNet-50 and Vision
Transformer (ViT) were compared for classifying oral
cancer and leukoplakia, with and without MixUp aug-
mentation. MixUp generates new data by linearly
combining images and labels, enhancing generaliza-
tion and reducing overfitting. Results showed that
MixUp improved accuracy for both models, with ViT
outperforming ResNet-50, especially when MixUp
was applied (Table 3). These findings highlight the
effectiveness of combining ViT and MixUp for this
task.
4.4 MSRCST Workflow
MSRCST generates randomly cropped images at
multiple scales (e.g., 240×250, 420×300 pixels) from
high-resolution images, ensuring a minimum side
length of 224 pixels and maintaining aspect ratios be-
tween 3:4 and 4:3. A teacher model (ViT-B 224) cal-
culates confidence scores for these patches, and high-
confidence patches are assigned pseudo-labels to cre-
ate a new dataset. This iterative process improves
model accuracy over multiple training cycles (Table
5). Experiments varying the number of cropped im-
ages (8, 12, 18) and selected patches (top 1, 2, 3, 6)
revealed that cropping 12 images and selecting the top
1 patch achieved the best accuracy of 71.71%.
4.5 Comparison with Other Methods
MSRCST was compared with two baseline methods:
1. Resize Method: Resizes all images to 224×224
pixels, ensuring full coverage but losing fine de-
tails.
2. Random Method: Uses all cropped patches with-
out confidence-based selection.
Using MixUp, the Resize method achieved 65.87%
accuracy, while the Random method reached 68.42%.
MSRCST outperformed both, achieving 71.71% ac-
curacy by focusing on high-confidence patches and
preserving diagnostic information.
4.6 MSRCST Verification Results
MSRCST consistently achieved high accuracy across
all conditions, demonstrating superior performance
compared to other methods (Table 4). Without
MixUp, MSRCST maintained a high accuracy of
69.62%, showing lower variability across folds.
Cropping 12 images and selecting the top 1 patch
achieved the best accuracy of 71.71%, while select-
ing six patches resulted in reduced accuracy. In mod-
els without MixUp, cropping 12 images and selecting
six patches yielded the best performance (69.00%).
4.7 Confusion Matrix Comparison
To evaluate classification performance, confusion ma-
trices for the MSRCST and Resize methods were an-
alyzed in terms of False Positives (FP), False Nega-
tives (FN), True Positives (TP), and True Negatives
(TN) (Figure 6). The Resize method had high FPs
and FNs, often misclassifying oral cancer as leuko-
plakia and vice versa, leading to lower TPs and TNs.
In contrast, MSRCST significantly reduced FPs and
FNs, achieving higher TPs and TNs. Cropping 12
images and selecting the top 1 patch resulted in the
highest accuracy (71.71%) with minimal misclassifi-
cation.
These results demonstrate that MSRCST effec-
tively reduces errors by focusing on diagnostically
relevant regions. By leveraging multi-scale cropping
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
784

Table 3: Model Performance Comparison with and without MixUp. This table compares the classification accuracy of
the ResNet-50 and ViT models with and without MixUp across different groups, and the weighted average is calculated and
presented.
Model MixUp Group 1 Group 2 Group 3 Group 4 Average
ResNet50 × 58.92% 73.64% 47.30% 67.51% 61.79%
ResNet50 ◦ 67.63% 76.57% 50.62% 69.20% 65.97%
ViT-B 224 × 69.29% 76.15% 41.08% 74.26% 65.13%
ViT-B 224 ◦ 78.42% 66.11% 60.17% 70.46% 68.79%
Table 4: Comparison of Classification Accuracy Across Methods. This table compares the classification accuracy of
MSRCST, Resize, and Random methods, both with (◦) and without (×) MixUp augmentation, using the weighted average
across each group.
Method MixUp Group 1 Group 2 Group 3 Group 4 Average
MSRCST × 63.48% 76.56% 64.32% 74.26% 69.62%
MSRCST ◦ 69.29% 86.61% 62.66% 68.35% 71.71%
Resize × 69.29% 76.15% 41.08% 74.26% 65.13%
Resize ◦ 78.42% 66.11% 60.17% 70.46% 68.79%
Random × 74.69% 71.97% 56.02% 72.15% 68.69%
Random ◦ 60.17% 73.64% 51.45% 69.20% 63.57%
Table 5: Evaluation of Confidence under Cropped Im-
age Count and MixUp Conditions. This table evaluates
the impact of the number of cropped images and the number
of selected patches on model accuracy under MixUp condi-
tions. It also compares the results with and without MixUp
when cropping 12 images.
Cropped Images MixUp
Top Confidence
Top 1 Top 2 Top 3 Top 6
8 Crops ◦ 69.10% 64.93% 70.14% 65.45%
12 Crops ◦ 71.71% 68.16% 66.59% 68.68%
18 Crops ◦ 69.63% 70.25% 67.85% 68.05%
12 Crops × 69.62% 68.27% 66.18% 69.00%
and confidence-based selection, MSRCST enhances
the model’s ability to distinguish oral cancer from
leukoplakia, improving diagnostic accuracy and reli-
ability.
5 DISCUSSION
In this study, we aimed to improve the classification
accuracy of oral cancer and leukoplakia using high-
resolution images and advanced deep learning tech-
niques. The results provide valuable insights into the
application of modern neural network architectures
and data augmentation strategies in medical imaging.
First, the Vision Transformer (ViT) demonstrated
superior performance compared to ResNet-50, pri-
marily due to its ability to capture both global and
local features using a self-attention mechanism. This
capability is particularly beneficial in medical imag-
ing tasks, where subtle differences between patho-
logical conditions are critical for accurate diagnosis.
ViT’s effectiveness highlights its potential for com-
plex medical applications requiring precise differenti-
ation.
Second, the MixUp data augmentation technique
improved classification performance for both ViT and
ResNet-50, with a greater impact observed in ViT. By
generating synthetic training samples through linear
combinations of images and labels, MixUp reduced
overfitting and enhanced model generalization. This
underscores the importance of robust data augmenta-
tion methods, especially when working with limited
datasets, as is common in medical research.
The proposed Multi-Scale Random Crop Self-
Training (MSRCST) method significantly outper-
formed the conventional Resize and Random meth-
ods. The Resize method, which downscales high-
resolution images to a standard size (e.g., 224×224
pixels), often loses critical diagnostic details, lead-
ing to misclassification, particularly for cases with
ambiguous lesion boundaries. The Random method,
which indiscriminately includes all cropped patches,
introduces noise by incorporating irrelevant regions.
In contrast, MSRCST preserves high-resolution in-
formation and focuses on diagnostically relevant re-
gions using confidence-based patch selection. This
approach reduces noise and improves classification
accuracy by ensuring that the model learns from the
Classification of Oral Cancer and Leukoplakia Using Oral Images and Deep Learning with Multi-Scale Random Crop Self-Training
785

Table 6: Confusion Matrices for Different Methods. Each subtable shows the confusion matrix for a different method of
image processing. Here, OC stands for Oral Cancer, LK for Leukoplakia, Pred for Predicted, and Act for Actual. The
four methods compared include different cut-out and adoption settings as well as the Resize method.
Pred OC Pred LK
Act OC 448 119
Act LK 152 239
(a) MSRCST (12 crops, Top 1)
Pred OC Pred LK
Act OC 425 142
Act LK 143 248
(b) MSRCST (18 crops, Top 2)
Pred OC Pred LK
Act OC 430 137
Act LK 149 242
(c) MSRCST (8 crops, Top 3)
Pred OC Pred LK
Act OC 461 106
Act LK 193 198
(d) ViT-Base 224 Resize
most relevant features.
Experiments revealed that generating 12 cropped
patches per image and selecting the top 1 or 2 patches
based on confidence yielded the highest accuracy
of 71.71%. This result indicates that focusing on
high-quality training samples improves learning effi-
ciency, while including excessive patches may intro-
duce noise and negatively impact accuracy. Notably,
MSRCST achieved a classification accuracy compa-
rable to the reported 70% accuracy of specialists di-
agnosing from images alone. However, further im-
provement is necessary to reach the higher accuracy
of in-person clinical diagnosis.
An important consideration is the potential impact
of data imbalance. The dataset used in this study
contained more images of oral cancer than leuko-
plakia, which could have biased the model. Nev-
ertheless, MSRCST effectively mitigated this imbal-
ance through its confidence-based selection process,
achieving balanced classification, as evidenced by
improved confusion matrices. This result suggests
that MSRCST prevents over-prediction of any single
class, ensuring robust performance across both condi-
tions.
The findings of this study hold significant clin-
ical implications. Improved classification models
can serve as valuable tools for non-specialist medi-
cal practitioners, enabling early detection and treat-
ment initiation for oral cancer and leukoplakia. Early
diagnosis not only prevents disease progression but
also enhances treatment success rates and improves
patients’ quality of life.
Future work should focus on expanding datasets
to include more cases and diverse imaging conditions.
This would improve model robustness and general-
ization. Additionally, integrating advanced data aug-
mentation methods and exploring novel model archi-
tectures could further enhance classification perfor-
mance. For example, combining imaging data with
clinical metadata may provide a more comprehensive
diagnostic approach. While specific methods and ar-
chitectures may evolve, the development of improved
strategies will likely address current limitations and
achieve higher diagnostic accuracy.
In conclusion, the combination of advanced deep
learning architectures such as the Vision Transformer,
effective data augmentation techniques like MixUp,
and the proposed MSRCST method significantly en-
hances the classification accuracy of oral cancer and
leukoplakia using high-resolution images. By focus-
ing on diagnostically relevant regions and reducing
noise, MSRCST provides a promising approach for
medical imaging tasks. Although further improve-
ments are needed for practical clinical applications,
this method represents a significant step toward en-
hancing early detection and treatment, ultimately im-
proving patient outcomes.
6 CONCLUSION
This paper has proposed Multi-Scale Random Crop
Self-Training (MSRCST) for classifying oral can-
cers and leukoplakia using images acquired by our
dermoscope. MSRCST comprises three key modules:
• Multi-Scale Random Crop: Extracts image
patches at various scales from high-resolution im-
ages, preserving both local details and global con-
textual information essential for accurate classifi-
cation.
• Selection Based on Confidence: Employs a
teacher model to assign confidence scores to each
cropped patch, selecting only those with high con-
fidence for further training. This ensures the
model focuses on diagnostically relevant features.
• Iteration of Self-Training: Iteratively retrains
the model using the selected high-confidence,
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
786

pseudo-labeled data, progressively enhancing ac-
curacy.
In our experiments, we applied MSRCST to clas-
sify images of oral cancer and leukoplakia. When
combined with MixUp data augmentation, MSRCST
achieved an average classification accuracy of
71.71%, outperforming traditional resizing and ran-
dom cropping methods. Additionally, it effectively
reduced misclassification rates, as demonstrated by
improved confusion matrices, thereby enhancing di-
agnostic reliability.
These results demonstrate that MSRCST success-
fully leverages high-resolution image data and semi-
supervised learning techniques to improve model per-
formance in medical image classification tasks. While
the study is limited by the dataset’s size and diversity,
future work will focus on expanding the dataset and
exploring additional techniques to further improve ac-
curacy and robustness.
REFERENCES
A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn,
X. Zhai, T. Unterthiner, M. Dehghani, M. Minderer,
G. Heigold, S. Gelly, J. Uszkoreit, and N. Houlsby. An
image is worth 16x16 words: Transformers for image
recognition at scale, 2021. URL https://arxiv.org/abs/
2010.11929.
K. He, X. Zhang, S. Ren, and J. Sun. Deep residual learning
for image recognition, 2015. URL https://arxiv.org/
abs/1512.03385.
Japan Society for Oral Cancer Elimination. What is oral
cancer? https://www.oralcancer.jp/2005p1/. [Ac-
cessed: 2024-10-22].
A. Krizhevsky, I. Sutskever, and G. E. Hinton. Ima-
genet classification with deep convolutional neural
networks. In Advances in Neural Information
Processing Systems, volume 25, 2012. URL https:
//proceedings.neurips.cc/paper files/paper/2012/file/
c399862d3b9d6b76c8436e924a68c45b-Paper.pdf.
D.-H. Lee. Pseudo-label : The simple and efficient semi-
supervised learning method for deep neural networks.
2013. URL https://api.semanticscholar.org/CorpusID:
18507866.
G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio,
F. Ciompi, M. Ghafoorian, J. A. van der Laak, B. van
Ginneken, and C. I. S
´
anchez. A survey on deep learn-
ing in medical image analysis. Medical Image Anal-
ysis, 42:60–88, Dec. 2017. ISSN 1361-8415. doi:
10.1016/j.media.2017.07.005. URL http://dx.doi.org/
10.1016/j.media.2017.07.005.
National Cancer Center Japan. Cancer statistics: Oral
and pharyngeal cancer. https://ganjoho.jp/reg stat/
statistics/stat/cancer/3 oral.html. [Accessed: 2024-
10-22].
T. Nomura, S. Matsubara, Y. Ro, A. Katakura, N. Takano,
T. Shibahara, and S. Seta. Usefulness of vital stain-
ing with iodine solution in resection of early tongue
carcinoma. Journal of The Japanese Stomatologi-
cal Society, 57(3):297–302, 2008. doi: 10.11277/
stomatology1952.57.297.
P. Rajpurkar, J. Irvin, K. Zhu, B. Yang, H. Mehta, T. Duan,
D. Ding, A. Bagul, C. Langlotz, K. Shpanskaya, M. P.
Lungren, and A. Y. Ng. Chexnet: Radiologist-level
pneumonia detection on chest x-rays with deep learn-
ing, 2017. URL https://arxiv.org/abs/1711.05225.
T. Shibahara, N. Yamamoto, T. Yakushiji, T. Nomura,
R. Sekine, K. Muramatsu, and H. Ohata. Narrow-band
imaging system with magnifying endoscopy for early
oral cancer. The Bulletin of Tokyo Dental College, 55
(2):87–94, 2014. doi: 10.2209/tdcpublication.55.87.
A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones,
A. N. Gomez, L. Kaiser, and I. Polosukhin. Attention
is all you need, 2017. URL https://arxiv.org/abs/1706.
03762.
Q. Xie, M.-T. Luong, E. Hovy, and Q. V. Le. Self-training
with noisy student improves imagenet classification,
2020. URL https://arxiv.org/abs/1911.04252.
H. Zhang, M. Cisse, Y. N. Dauphin, and D. Lopez-Paz.
mixup: Beyond empirical risk minimization, 2018.
URL https://arxiv.org/abs/1710.09412.
Classification of Oral Cancer and Leukoplakia Using Oral Images and Deep Learning with Multi-Scale Random Crop Self-Training
787
