
Breast Density Estimation in Mammograms Using Unsupervised
Image Segmentation
Khaldoon Alhusari and Salam Dhou
Department of Computer Science and Engineering, American University of Sharjah, Sharjah, U.A.E.
Keywords: Breast Cancer, Breast Density, Mammograms, Unsupervised Learning, Segmentation.
Abstract: Breast cancer is very common, and early detection through mammography is paramount. Breast density, a
strong risk factor for breast cancer, can be estimated from mammograms. Current density estimation methods
can be subjective, labor-intensive, and proprietary. This work proposes a framework for breast density
estimation based on the unsupervised segmentation of mammograms. A state-of-the-art unsupervised image
segmentation algorithm is adopted for the purpose of breast density segmentation. Mammographic percent
density is estimated through a process of arithmetic division. The percentages are then discretized into
qualitative assessments of density (“Fatty” and “Dense”) using a thresholding approach. Evaluation reveals
robust segmentation at the pixel-level with silhouette scores averaging 0.95 and significant unsupervised
labeling quality at the per-image level with silhouette scores averaging 0.61. The proposed framework is
highly adaptable, generalizable, and non-subjective, and has the potential to be a beneficial support tool for
radiologists.
1 INTRODUCTION
Breast cancer remains a formidable global health
challenge, constituting 11.7% of all cancer cases with
alarming mortality rates (Sung et al., 2021). The
significance of early detection is underscored by its
potential to mitigate the risks associated with this
prevalent malignancy. Mammographic screening,
particularly through mammograms, stands out as a
pivotal diagnostic tool due to its efficacy in
identifying tumors before clinical symptoms
manifest. However, the efficacy of mammography is
intricately linked to breast density, a parameter
determined by the proportion of radio-dense,
fibroglandular tissue (Kallenberg et al., 2016). Higher
breast density complicates tumor identification,
particularly in cases where the mammogram appears
highly dense, making early detection less reliable
(Wengert et al., 2019). Despite its efficacy,
mammography introduces a critical concern—
radiation exposure. The risk, though minimal, is not
negligible, and it is exacerbated in women with larger
breasts of a higher density (Dhou et al., 2022).
The interpretative aspect of mammography
introduces another layer of complexity, especially
concerning breast density assessments. Radiologists
exhibit variability in determining breast density, with
notable subjectivity in cases of highly dense breasts
(Sprague et al., 2016). This subjectivity can lead to
missed cancer diagnoses and biased treatment
decisions, as evidenced by the tendency to opt for
more invasive procedures for extremely dense breasts
(Nazari & Mukherjee, 2018).
There are various methods of breast density
estimation in the literature. In practice, the most
common is a visual assessment method known as the
Breast Imaging-Reporting and Data System (BI-
RADS) (Sickles, EA, D’Orsi CJ, Bassett LW, 2013),
which classifies breasts into four qualitative density
categories. Other visual systems of measurement
exist, such as the Wolfe Classification (Wolfe, 1976),
the Tabár Classification (Gram et al., 1997), and the
Visual Analogue Scale (VAS) (Nahler, 2009). There
are also software-based methods for breast density
estimation, some of which are semi-automatic such as
the Cumulus software (Byng et al., 1994), and some
of which are fully automatic such as the Quantra
software (Hartman et al., 2008).
In addition, there are studies that use machine
learning to solve the problem of breast density
estimation. It is worth noting that, using
mammography, AI has been successful in performing
several vital tasks such as tumor detection,
Alhusari, K. and Dhou, S.
Breast Density Estimation in Mammograms Using Unsupervised Image Segmentation.
DOI: 10.5220/0013297500003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 3: VISAPP, pages
691-698
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
691

classification, image improvement and breast density
estimation (Dhou et al., 2024). In a relevant work,
(Arefan et al., 2015), nine statistical features are
extracted from preprocessed mammograms and fed
into a two-layer feed-forward neural network,
achieving high classification accuracy. The work in
(Saffari et al., 2020) employs a conditional generative
adversarial network (cGAN) in combination with a
convolutional neural network (CNN) to classify
mammograms into one of four density categories, and
achieves high agreement with expert assessment.
Further, there are methods that rely on the
segmentation of the mammogram to produce a
percentage estimate of mammographic density. These
can be area-based, relying on the 2-dimensional
appearance of mammograms (e.g., thresholding and
clustering), or volume-based, attempting to quantify
the volume of density within a breast by estimating
depth. Notable works include (Kallenberg et al.,
2016), which uses a convolutional sparse autoencoder
(CSAE) with softmax regression, and (Gudhe et al.,
2022), which employs a multitask deep learning
model that utilizes multilevel dilated residual blocks
and parallel dilated convolutions to enhance feature
extraction. Both of these works achieve significant
correlation with expert assessment.
Each of the detailed breast density estimation
methods has some merits, but is also liable to flaws.
Visual methods like BI-RADS are widely adopted but
subjective and labor-intensive. Breast density
software are convenient but present challenges in
relation to their proprietary nature, and are often
inconsistent with one another. Machine learning
methods show promise but rely on subjective label
data and often focus on BI-RADS classification rather
than quantitative estimation. Segmentation-based
approaches offer quantitative estimates but face
challenges, with some area-based methods being
unsupervised but primitive, volumetric methods
being complex and inconvenient, and sophisticated
methods having to rely on supervised techniques and
hand-annotated data.
This work aims to present a robust and practical
solution to the problem of subjectivity in breast
density estimation. It proposes a framework that
makes use of deep-learning-based unsupervised
segmentation followed by arithmetic division to
achieve quantitative percentage density estimates. As
part of the framework, mammogram preprocessing
methods for breast reorientation, artifact and noise
removal, and region of interest (ROI) extraction are
presented. A state-of-the-art unsupervised
segmentation algorithm based on a CNN whose loss
function combines similarity and continuity is
adopted and tuned for the purpose of breast density
segmentation. An arithmetic division approach for
percentage density estimation is employed, and
estimated densities are then discretized into binary
(“Fatty” and “Dense”) classes through a thresholding
approach. Experimental results show that the
proposed framework has the potential to be of benefit
to radiologists in a clinical setting as a support tool
for quantifying breast density.
2 METHODOLOGY
2.1 Dataset
This work uses the INbreast dataset (Moreira et al.,
2012), which is a public mammography dataset. It has
a total of 410 images belonging to 115 patients. Of
those images, 203 are CC images, and 206 are MLO
images. Following the image selection and
preprocessing phases, 306 images are left, and they
can be attributed to 111 patients. 104 of those patients
have at least two images remaining, while 41 have
exactly four. Only 7 patients end up with a single
image.
In terms of labels, 210 of the 306 images are
classified by experts as fatty and assigned either the
BIRADS I or the BIRADS II label. The other 96 are
classified as dense and assigned either the BIRADS
III or the BIRADS IV label. The distribution of
individual images and patients is shown in Table 9.
104 images are assigned to the BIRADS I class, 106
are assigned the BIRADS II class, 73 are assigned the
BIRADS III class, and 23 are assigned the BIRADS
IV class. Thus, the vast majority of the images are
assigned to the first and second BIRADS classes, and
only a small percentage is assigned to the fourth
BIRADS class.
The images of the INbreast dataset come in the
DICOM format, meaning that they had to be
converted to the JPEG format prior to any processing.
The resultant images each had one of two sizes, 2560
× 3328 pixels or 3328 × 4084 pixels. The images had
very little noise and very few artifacts, but not few
enough to make the artifact removal process
unnecessary.
Tumors and dense tissue look very similar within
a mammogram (Birdwell, 2009). Since this is the
case, only negative mammograms (non-malignant)
are selected and used in this work, as is a common
practice in the medical field. Both mammogram
views—Mediolateral Oblique (MLO) and
Craniocaudal (CC)—are incorporated.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
692

2.2 Image Preprocessing
To prepare mammograms for segmentation and
density estimation, three image preprocessing steps
are necessary. First, the mammograms, within which
the breast can be left or right-facing, must be
reoriented such that all breasts face the same
direction. In this work, all mammograms are
reoriented such that the breasts are right-facing.
Inspired by the work in (Dehghani & Dezfooli, 2011),
the mammograms are binarized and split horizontally
down the middle. The pixel sums on each side are
calculated, and the side with the higher sum is the one
containing the breast—this yields the current
orientation of the breast. The mammogram is then
flipped across the y-axis if the breast within it is
determined to be left facing.
Following reorientation, noise and artifacts within
the mammogram must be removed to facilitate ROI
extraction. To achieve this, the input mammogram is
binarized again, and morphological closing is applied
to make sure each apparent island, breast tissue
included, is fully connected. Then, the largest contour
in the binary image, presumably the breast, is used as
a mask and applied to the original mammogram
through a bitwise AND operation. This process leaves
a mammogram with all artifacts removed without
making any changes to the original shape of the breast
or the size of the image.
Lastly, ROIs can be extracted from the central
region of the breast, since that is the area most
indicative of differences between distinct breast
density categories (Li et al., 2004). A fixed size of
128x128 pixels is selected, which resulted in better
classification accuracy when compared to a size of
64x64 pixels in (Saffari et al., 2020). This
enhancement is likely due to the improved feature
representation achieved by including a larger portion
of the central region. Figure 1 shows samples of the
extracted ROIs for this work.
Figure 1: Sample ROIs extracted from mammograms.
2.3 Unsupervised Segmentation
The unsupervised segmentation model proposed in
(Kim et al., 2020) is adopted in this work. In it, the
authors built a CNN for general-purpose,
unsupervised image segmentation. The CNN consists
of a modifiable number of convolutional layers (with
a minimum of two), each of which applies a 2D
convolution operation to the input data, and batch
normalization layers, which normalize the activations
of the previous layer. Between each pair of
convolutional and batch normalization layers, there is
a ReLU activation function, which introduces non-
linearity to the model. The model can be used on input
images to produce segmentation masks that divide the
images into distinct regions based on the similarity of
pixels within the images. This approach aims to
minimize a combination of similarity loss and spatial
continuity loss in order to find a suitable solution for
assigning labels to the different regions of the image.
The algorithm is reiterated until either a specified
minimum number of labels or a specified number of
iterations is reached. Reported experimental results
show that this approach is highly capable when it
comes to image segmentation.
As can be seen in Figure 2, the algorithm works
through a process of back propagation. In the forward
pass, after an image is passed through the CNN, a
response map is produced. The response map holds
all of the potential labels for each pixel within the
image, as well as the probability or likelihood that
each of those labels is the correct one. Argmax
classification is then applied to the response map.
This essentially selects the label with the highest
probability for each pixel within the response map,
thereby producing the cluster labels for the image.
Following this, the loss can be computed.
The loss function, as noted previously, combines
similarity and spatial continuity losses. The two
constraints are as follows:
i. Similarity constraint: pixels with similar
characteristics should be assigned the same label.
ii. Continuity constraint: neighboring pixels should
be assigned to the same label.
With those constraints in mind, the loss equation
is as follows (Kim et al., 2020):
,
(1)
where
denotes similarity loss,
denotes
continuity loss,
denotes the normalized response
map,
denotes the cluster labels, and is a weight
for balancing the two constraints.
For the similarity constraint, cross entropy is
used. This is the case for the original algorithm, as
well as the adopted algorithm, since testing proved it
to be the best performing similarity loss function for
the purposes of this work. The cross entropy loss
equation employed is as follows (Kim et al., 2020):
Breast Density Estimation in Mammograms Using Unsupervised Image Segmentation
693
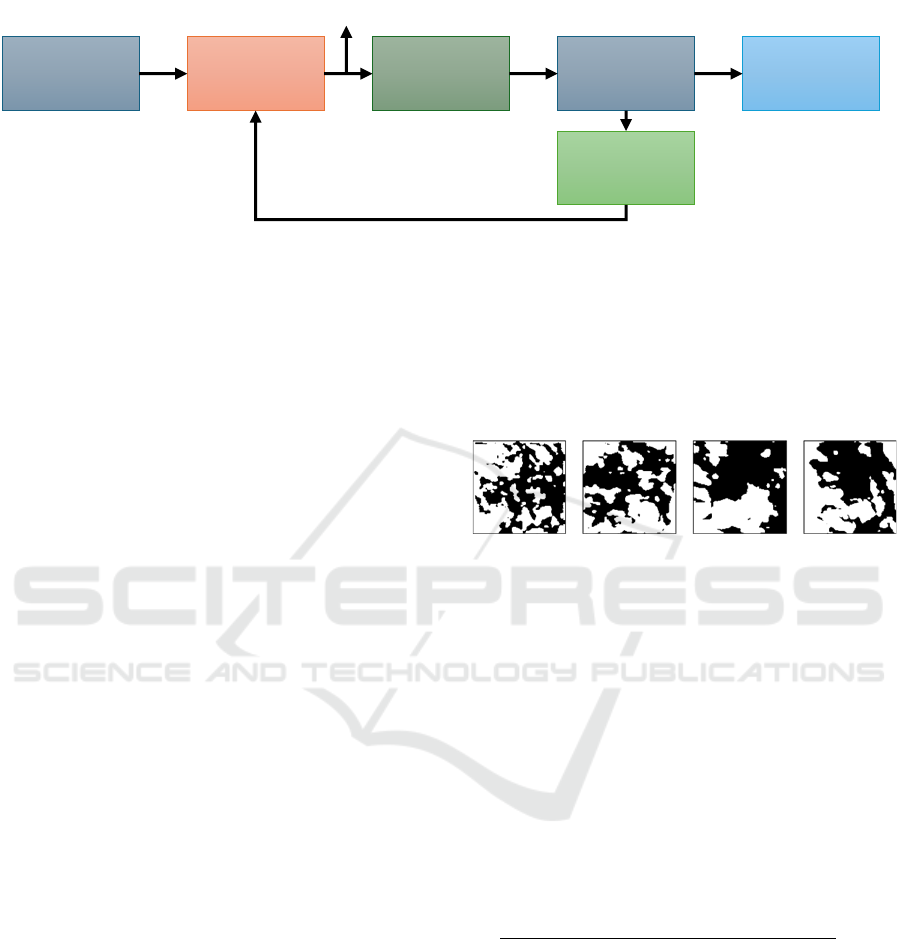
Figure 2: Flow of the segmentation algorithm adopted in this work (Kim et al., 2020). In the forward pass, the CNN processes
the input image to produce a response map. Argmax classification finds the highest probability label for each pixel. The model
then calculates loss, computes gradients, and updates parameters through the backward pass.
,
(2)
where N is the number of pixels in the response map,
q is the number of cluster labels for each pixel in the
response map, and δ is a function of t, where δ(t) is
represented by the following equation (Kim et al.,
2020):
(3)
For the continuity constraint, L1 loss, also known
as the mean absolute error (MAE) loss, is used by the
algorithm. The L1 loss equation is as follows (Kim et
al., 2020):
(4)
For Equation 4, W and H are the width and height
of the input image, and
is the pixel value at (ε,η)
in the response map
. The equation is applied for
the vertical and the horizontal components of the
input image.
Following the computation of the loss function, a
backward pass is initiated to compute gradients of the
loss with respect to the model's weights and biases.
The algorithm also makes use of a Stochastic
Gradient Descent (SGD) optimizer to update the
CNN parameters based on the computed gradients,
and the original image is passed through the CNN—
now with newly updated parameters—once again.
The whole process repeats until either the maximum
number of iterations (1000 by default) is reached, or
the CNN reaches convergence at the minimum
number of labels (set to 2). Samples of the resulting
segmentation masks are shown in Figure 3.
Figure 3: Segmentation results of the ROI samples shown
in Figure 1.
2.4 Breast Density Estimation
For the task of breast density estimation, continuous
percentage density estimates can be computed given
the segmented image masks using arithmetic division.
In essence, the segmented image is a binary image
with two regions (clusters). One of those regions is
dense, and the other is fatty. To calculate the
percentage density, the number of pixels in the dense
region is counted, divided by the total number of
pixels in the image, and multiplied by 100.
Mathematically, the equation is as follows:
(5)
The resultant density estimates are quantitative,
and can be discretized to represent qualitative labels.
To do this, a threshold is calculated based on the mean
density and the standard deviation across all images
in the dataset. Applying this threshold, the result is a
list of qualitative density labels, each pertaining to a
particular image. The qualitative labels produced in
this process are utilized in the clustering quality
evaluation described in Section 2.5 below.
CNN
Input
Image
Argmax
Classification
Cluster Labels
Loss
Calculation
Back Propagation
Segmented
Image
Response Map
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
694

2.5 Performance Evaluation
The evaluation process for this work comprises two
steps: an evaluation of the segmentation quality at the
pixel level and an assessment of the unsupervised
clustering quality at the per-image level. The metrics
used here are as follows: the silhouette coefficient
(SC), which measures cohesion and separation among
clusters using the following equation:
(6)
where A is the mean distance between a sample and
all other points in the same cluster, and B is the mean
distance between a sample and all other points in the
nearest cluster to which the sample does not belong;
the Within-Cluster Sum of Squares (WCSS), which
quantifies the compactness of clusters using the
following equation:
(7)
where k is the number of clusters, C
i
is the i
th
cluster,
x is a data point in the cluster, u
i
is the centroid of the
i
th
cluster, and “||” denotes the Euclidean norm; the
Davies-Bouldin (DB) score, which assesses average
similarity between clusters using the following
equation:
(8)
where k is the number of clusters, S
i
is the average
distance between data points in cluster i and the
centroid of cluster i, with Sj being the cluster j
equivalent, and D
ij
is the distance between the
centroids of clusters i and j; and the Calinski-
Harabasz (CH) index, which evaluates the ratio of
between-cluster variance to within-cluster variance
using the following equation:
(9)
where B is the between-cluster variance, W is the
within-cluster variance, N is the total number of data
points, and k is the number of clusters. To facilitate
the evaluation of the framework’s clustering ability at
the per-image level, nine features are extracted from
the images: mean luminance, standard deviation,
entropy, intensity ranges, 25
th
, 50
th
, and 75
th
percentiles, skewness, and kurtosis.
3 RESULTS
This section presents and discusses the results
following the evaluation of the framework described
in this work. First, an evaluation of the segmentation
is provided. Then, an evaluation of the framework’s
unsupervised labeling ability is presented. Lastly, a
discussion of the results is presented.
3.1 Segmentation Evaluation
The quality of the segmentation is evaluated at the
pixel level through the use of four metrics: SC,
WCSS, DB, and CH. This evaluation is performed
separately for the CC and MLO subsets of the dataset.
The results are shown in Table 1.
Table 1: Segmentation evaluation for CC and MLO subsets
of the INbreast dataset.
Metric\Subset
CC
MLO
SC
0.9491
0.9505
WCSS
3,224,218.76
2,908,076.60
DB
0.0725
0.0697
CH
19,471.41
13,613.63
The SC Scores average around 0.9491 for the CC
subset and range from 0.8974 to 0.9907, suggesting
notable cluster separation throughout. The WCSS
evaluation reveals an average of 3,224,218.76, with a
range between 150,933.91 and 14,200,223.75. DB
Scores have an average of 0.0725 for the CC subset,
and range from 0.0119 to 0.1813, suggesting
generally well-defined clusters. The CH Indices have
an average of 19,471.41, and range between 6.87 and
896,492.27.
In the MLO subset, SC Scores demonstrate an
average of 0.9505, and range between 0.8851 and
0.9929, showing effective segmentation. The WCSS
scores for the MLO subset have an average of
2,908,076.60, and a range from 98,592.53 to
12,226,390.39. DB Scores present an average value
of 0.0697, and a range between 0.0096 and 0.1391,
implying robust segmentation overall. The CH
Indices yield an average index value of 13,613.63,
and a range between 0.60 and 42,226.4.
Breast Density Estimation in Mammograms Using Unsupervised Image Segmentation
695
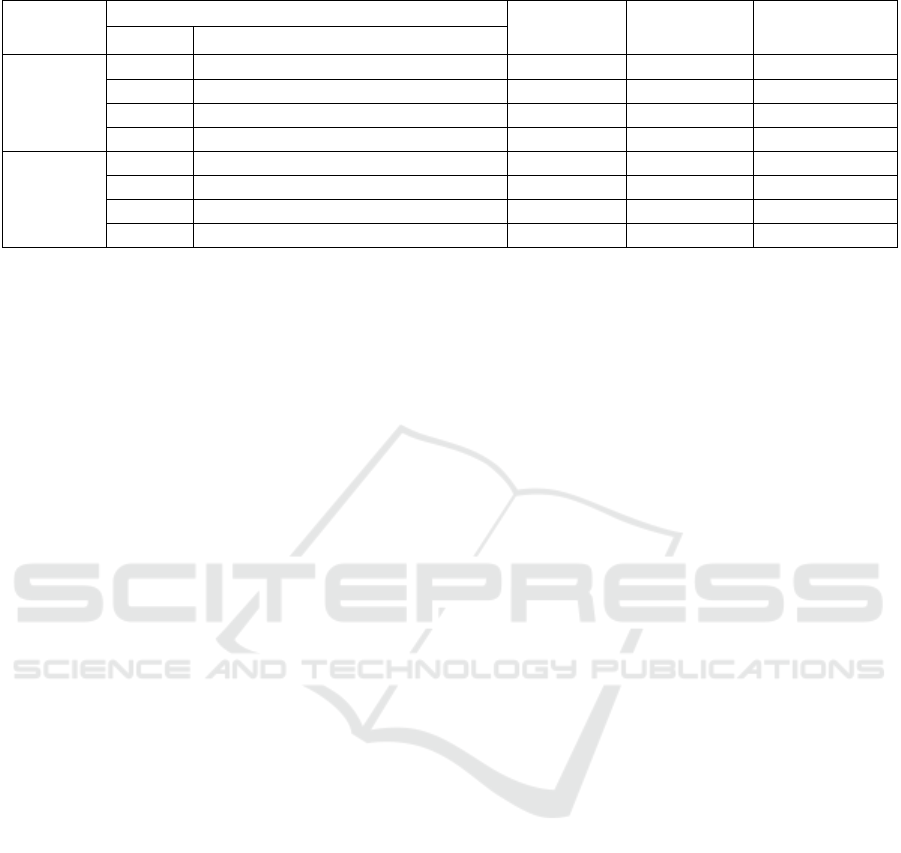
Table 2: Clustering quality evaluation for CC and MLO subsets of the INbreast dataset using a varying number of features.
Subset
Features
Silhouette
Davies-
Bouldin
Calinski-
Harabasz
Number
Set
CC
1
25
th
Percentile
0.9101
0.2519
1383.58
2
25
th
Percentile, Luminance
0.8698
0.3245
762.69
3
25
th
and 50
th
Percentiles, Luminance
0.8469
0.3795
549.74
9
All
0.6238
0.6889
104.15
MLO
1
Kurtosis
0.9416
0.0787
1806.60
2
Skewness, Kurtosis
0.9036
0.128
1016.62
3
Skewness, Kurtosis, 50
th
Percentile
0.8437
0.2260
772.06
9
All
0.5977
0.6373
199.47
3.2 Clustering Quality Evaluation
In this section, the clustering quality, or the quality of
the unsupervised labeling of images as fatty or dense,
is evaluated. This is done with metrics that do not rely
on ground truth, and instead measure the quality of
and separation between clusters: the SC, the DB
Score, and the CH Index. The metrics were computed
separately for CC and MLO images based on
extracted statistical features—mean luminance,
standard deviation, statistical entropy, pixel intensity
ranges (the maximum minus the minimum for each
image), the 25th, 50th, and 75th percentiles,
skewness, and kurtosis—after MinMax scaling. The
results are shown in Table 2. In the context of breast
density estimation, the SC indicates well-defined
density categories, the CH index shows the
distinctness and compactness of clusters, and the DB
score measures the separation and cohesion of these
clusters. Higher SC and CH values and a lower DB
score indicate better clustering quality.
Combining all features, the resulting SC score for
CC images was 0.6238, which indicates notable
cohesion and separation between clusters. The DB
score was 0.6889, and the CH Index was 104.15, both
of which suggest a reasonable degree of separation
between clusters. Using only the 25th percentile to
represent the images results in the highest SC score
(0.9101). Using mean luminance and the 25th
percentile is also representative, resulting in a SC
score of 0.8698. When using three features,
combining the mean luminance and the 25th
percentile with the 50th percentile is the most
effective for CC images, resulting in a SC score of
0.8469.
For the MLO subset of the dataset, the
combination of all features results in a SC score of
0.5977, a DB score of 0.6373, and a CH index of
199.47. This indicates a notable degree of separation
and cohesion between and among clusters. In contrast
to CC, the most representative feature for MLO is
kurtosis, and it results in a SC score of 0.9416.
Combining kurtosis with skewness results in a SC
score of 0.9036. For three features, the highest SC
score (0.8437) results from combining skewness,
kurtosis, and the 50th percentile.
3.3 Discussion
The segmentation evaluation shows strong pixel-
level performance for both CC and MLO images,
with high SC scores indicating good cluster cohesion
and separation. DB scores suggest effective
clustering, though WCSS and CH scores vary. For
unsupervised labeling of Fatty or Dense, key features
like the 25th percentile and mean luminance for CC,
and kurtosis and skewness for MLO, effectively
distinguish clusters. While adding more features
might introduce noise, combining all features still
achieves significant separation between Fatty and
Dense categories.
These results are promising and suggest that the
proposed framework can distinguish between Fatty
and Dense breasts in an unsupervised manner. This is
a non-subjective approach as it does not rely on expert
assessment. In contrast, the current state-of-the-art
methods exclusively rely on expert-assigned labels
for classification and hand-annotated markings for
segmentation. Additionally, the framework is highly
generalizable, given that it does not need to be trained
on a specific dataset, and it is also highly adaptable.
Furthermore, it is inexpensive and fully automatic,
which means that it is easily integrable into clinical
settings.
The novelty of this work lies in the
implementation of unsupervised image segmentation
to solve the problem of breast density estimation,
which has not been done before. Though the
segmentation algorithm is directly adopted with
limited changes, it is extensively tuned for the task
and integrated into a task-specific, scalable
framework, with automated image selection, breast
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
696

reorientation, artifact removal, ROI extraction, and
continuous and binary density estimation modules.
Numerical comparison of the results to other works in
the literature is not possible at this stage, as they
exclusively employ traditional classification metrics.
In the future, the agreement between the expert labels
and the framework’s binary labels will be measured,
enabling comparison to other works through
classification metrics.
4 CONCLUSION
This work introduced a framework for breast density
estimation through unsupervised segmentation of
mammographic images. It includes preprocessing
methods for breast reorientation, artifact and noise
removal, and ROI extraction. A state-of-the-art
segmentation algorithm was tuned for breast density
segmentation, and percentage density estimation was
performed using an arithmetic division approach.
Breast density was then discretized into two classes,
Fatty and Dense, via a thresholding approach. The
framework’s segmentation quality and unsupervised
labeling ability were evaluated, showing robust
performance. For segmentation at the pixel-level,
silhouette scores averaging 0.95 were achieved.
Further, for the unsupervised labeling of
mammograms, an average silhouette score of 0.61
was attained. This suggests the framework’s potential
as a support tool for radiologists in a clinical setting.
For future work, the framework’s agreement with
expert labels will be evaluated. Further, other datasets
will be used to test and verify the generalizability of
the framework. In addition, supplemental testing will
be conducted to determine if the framework can be
further refined, such as through the use of ROIs with
adaptive sizes rather than fixed sizes, or through the
employment of other unsupervised segmentation
algorithms. Moreover, to improve the error-handling
ability of the framework, a postprocessing procedure
will be implemented to reassign labels to incorrectly
classified images through the use of a confidence
metric.
REFERENCES
Arefan, D., Talebpour, A., Ahmadinejhad, N., & Asl, A. K.
(2015). Automatic breast density classification using
neural network. Journal of Instrumentation, 10(12).
https://doi.org/10.1088/1748-0221/10/12/T12002
Birdwell, R. L. (2009). The preponderance of evidence
supports computer-aided detection for screening
mammography. In Radiology (Vol. 253, Issue 1).
https://doi.org/10.1148/radiol.2531090611
Byng, J. W., Boyd, N. F., Fishell, E., Jong, R. A., & Yaffe,
M. J. (1994). The quantitative analysis of
mammographic densities. Physics in Medicine and
Biology, 39(10). https://doi.org/10.1088/0031-
9155/39/10/008
Dehghani, S., & Dezfooli, M. A. (2011). A Method For
Improve Preprocessing Images Mammography.
International Journal of Information and Education
Technology. https://doi.org/10.7763/ijiet.2011.v1.15
Dhou, S., Alhusari, K., & Alkhodari, M. (2024). Artificial
intelligence in mammography: advances and
challenges. In Artificial Intelligence and Image
Processing in Medical Imaging (pp. 83–114). Elsevier.
https://doi.org/10.1016/B978-0-323-95462-4.00004-2
Dhou, S., Dalah, E., AlGhafeer, R., Hamidu, A., &
Obaideen, A. (2022). Regression Analysis between the
Different Breast Dose Quantities Reported in Digital
Mammography and Patient Age, Breast Thickness, and
Acquisition Parameters. Journal of Imaging, 8(8), 211.
https://doi.org/10.3390/jimaging8080211
Gram, I. T., Funkhouser, E., & Tabár, L. (1997). The Tabar
classification of mammographic parenchymal patterns.
European Journal of Radiology, 24(2).
https://doi.org/10.1016/S0720-048X(96)01138-2
Gudhe, N. R., Behravan, H., Sudah, M., Okuma, H.,
Vanninen, R., Kosma, V. M., & Mannermaa, A. (2022).
Area-based breast percentage density estimation in
mammograms using weight-adaptive multitask
learning. Scientific Reports, 12(1).
https://doi.org/10.1038/s41598-022-16141-2
Hartman, K., Highnam, R., Warren, R., & Jackson, V.
(2008). Volumetric assessment of breast tissue
composition from FFDM images. Lecture Notes in
Computer Science (Including Subseries Lecture Notes
in Artificial Intelligence and Lecture Notes in
Bioinformatics), 5116 LNCS.
https://doi.org/10.1007/978-3-540-70538-3_5
Kallenberg, M., Petersen, K., Nielsen, M., Ng, A. Y., Diao,
P., Igel, C., Vachon, C. M., Holland, K., Winkel, R. R.,
Karssemeijer, N., & Lillholm, M. (2016). Unsupervised
Deep Learning Applied to Breast Density Segmentation
and Mammographic Risk Scoring. IEEE Transactions
on Medical Imaging, 35(5).
https://doi.org/10.1109/TMI.2016.2532122
Kim, W., Kanezaki, A., & Tanaka, M. (2020).
Unsupervised Learning of Image Segmentation Based
on Differentiable Feature Clustering. IEEE
Transactions on Image Processing, 29.
https://doi.org/10.1109/TIP.2020.3011269
Li, H., Giger, M. L., Huo, Z., Olopade, O. I., Lan, L.,
Weber, B. L., & Bonta, I. (2004). Computerized
analysis of mammographic parenchymal patterns for
assessing breast cancer risk: Effect of ROI size and
location. Medical Physics, 31(3).
https://doi.org/10.1118/1.1644514
Moreira, I. C., Amaral, I., Domingues, I., Cardoso, A.,
Cardoso, M. J., & Cardoso, J. S. (2012). INbreast:
Toward a Full-field Digital Mammographic Database.
Breast Density Estimation in Mammograms Using Unsupervised Image Segmentation
697

Academic Radiology, 19(2).
https://doi.org/10.1016/j.acra.2011.09.014
Nahler, G. (2009). visual analogue scale (VAS). In
Dictionary of Pharmaceutical Medicine.
https://doi.org/10.1007/978-3-211-89836-9_1450
Nazari, S. S., & Mukherjee, P. (2018). An overview of
mammographic density and its association with breast
cancer. In Breast Cancer (Vol. 25, Issue 3).
https://doi.org/10.1007/s12282-018-0857-5
Saffari, N., Rashwan, H. A., Abdel-Nasser, M., Singh, V.
K., Arenas, M., Mangina, E., Herrera, B., & Puig, D.
(2020). Fully automated breast density segmentation
and classification using deep learning. Diagnostics,
10(11). https://doi.org/10.3390/diagnostics10110988
Sickles, EA, D’Orsi CJ, Bassett LW, et al. (2013). ACR BI-
RADS® Mammography. In: ACR BI-RADS® Atlas,
Breast Imaging Reporting and Data System. Reston,
VA, American College of Radiology.
Sprague, B. L., Conant, E. F., Onega, T., Garcia, M. P.,
Beaber, E. F., Herschorn, S. D., Lehman, C. D.,
Tosteson, A. N. A., Lacson, R., Schnall, M. D., Kontos,
D., Haas, J. S., Weaver, D. L., & Barlow, W. E. (2016).
Variation in Mammographic Breast Density
Assessments among Radiologists in Clinical Practice:
A Multicenter Observational Study. Annals of Internal
Medicine, 165(7). https://doi.org/10.7326/M15-2934
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M.,
Soerjomataram, I., Jemal, A., & Bray, F. (2021). Global
Cancer Statistics 2020: GLOBOCAN Estimates of
Incidence and Mortality Worldwide for 36 Cancers in
185 Countries. CA: A Cancer Journal for Clinicians,
71(3). https://doi.org/10.3322/caac.21660
Wengert, G. J., Helbich, T. H., Leithner, D., Morris, E. A.,
Baltzer, P. A. T., & Pinker, K. (2019). Multimodality
Imaging of Breast Parenchymal Density and
Correlation with Risk Assessment. In Current Breast
Cancer Reports (Vol. 11, Issue 1).
https://doi.org/10.1007/s12609-019-0302-6
Wolfe, J. N. (1976). Breast patterns as an index of risk for
developing breast cancer. American Journal of
Roentgenology, 126(6).
https://doi.org/10.2214/ajr.126.6.1130
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
698
