
Domain Generalization Using Category Information
Independent of Domain Differences
Reiji Saito
a
and Kazuhiro Hotta
b
Meijo University, 1-501 Shiogamaguchi, Tempaku-ku, Nagoya 468-8502, Japan
Keywords:
Domain Generalization, Segmentation, DeepCCA, SQ-VAE.
Abstract:
Domain generalization is a technique aimed at enabling models to maintain high accuracy when applied to new
environments or datasets (unseen domains) that differ from the datasets used in training. Generally, the accu-
racy of models trained on a specific dataset (source domain) often decreases significantly when evaluated on
different datasets (target domain). This issue arises due to differences in domains caused by varying environ-
mental conditions such as imaging equipment and staining methods. Therefore, we undertook two initiatives
to perform segmentation that does not depend on domain differences. We propose a method that separates cat-
egory information independent of domain differences from the information specific to the source domain. By
using information independent of domain differences, our method enables learning the segmentation targets
(e.g., blood vessels and cell nuclei). Although we extract independent information of domain differences, this
cannot completely bridge the domain gap between training and test data. Therefore, we absorb the domain gap
using the quantum vectors in Stochastically Quantized Variational AutoEncoder (SQ-VAE). In experiments,
we evaluated our method on datasets for vascular segmentation and cell nucleus segmentation. Our methods
improved the accuracy compared to conventional methods.
1 INTRODUCTION
Semantic segmentation is a technique for classifying
images at the pixel level and is applied in various
fields such as medical imaging (J.Wang et al., 2020;
F.Milletari et al., 2016), autonomous driving (Y.Liu
et al., 2020), and cellular imaging (Furukawa and
Hotta, 2021; Shibuya and Hotta, 2020). Conventional
methods (P.Wang et al., 2023; B.Cheng et al., 2022)
are typically trained on specific datasets (source do-
mains) and evaluated on the same datasets. However,
these methods often perform poorly when evaluated
on different datasets (target domains) due to domain
shift. In medical segmentation, domain shift is par-
ticularly pronounced because images are captured in
various hospitals and clinical settings. Domain shift
occurs due to differences in imaging conditions, such
as imaging devices, lighting, and staining methods.
Ideally, accuracy should be maintained regardless of
the dataset used for evaluation. Addressing this do-
main shift and effectively extracting category infor-
mation that is independent of these differences is a
long-standing challenge in deep learning.
a
https://orcid.org/0009-0003-5197-8922
b
https://orcid.org/0000-0002-5675-8713
One common approach to solving the domain shift
problem is domain adaptation (DA). DA leverages la-
beled data from the source domain to adjust its distri-
bution to match the target domain, maximizing per-
formance on the target domain. However, this ap-
proach requires capturing and learning from target do-
main images, which can be time-consuming. Addi-
tionally, DA is only applicable to the specific target
domain images being trained, lacking generalizabil-
ity. Furthermore, in segmentation tasks, manual an-
notation is required, which can be a significant burden
for researchers.
Domain generalization (DG) has been proposed to
address the limitations of DA. DG leverages only the
source domain to extract features that are not specific
to it (e.g., cell nuclei and blood vessels), thereby mit-
igating domain shift when encountering unseen target
domains. Here, we focus on developing a model that
effectively generalizes across diverse medical imag-
ing conditions, enhancing robustness and adaptability
to varying environments. Research on DG (S.Choi
et al., 2021; X.Pan et al., 2018) has developed meth-
ods that eliminate domain-specific style information
from images and use content information for learning.
Specifically, these methods involve whitening style-
368
Saito, R. and Hotta, K.
Domain Generalization Using Category Information Independent of Domain Differences.
DOI: 10.5220/0013300300003905
In Proceedings of the 14th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2025), pages 368-376
ISBN: 978-989-758-730-6; ISSN: 2184-4313
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

specific features based on the correlation of feature
values, thereby retaining content information and im-
proving generalization performance. However, Wild-
Net (S.Lee et al., 2022) noted that style informa-
tion also contains essential features for semantic cat-
egory prediction, and addressing this issue has been
reported to improve accuracy.
To address DG without removing style infor-
mation, we have employed the following two ap-
proaches. First, we proposed a method to split fea-
ture maps into two parts: domain-invariant category
information and source domain-specific information.
Specifically, we divide the feature maps along the
channel dimension and use DeepCCA (G.Andrew
et al., 2013) to decorrelate these parts. DeepCCA
maximizes the correlation between two variables,
but we train it to make the correlation zero to ex-
tract source domain-specific information and domain-
invariant category information. We train one of the
split feature maps to represent domain-invariant cat-
egory information. Specifically, we train the feature
vectors of the same category, based on the ground
truth labels, to approach a learnable representative
vector. Since the two feature maps are decorrelated,
the other feature map becomes the source domain-
specific information. We use the obtained domain-
invariant category features for segmentation. Sec-
ond, although we extracted domain-invariant cate-
gory information, this alone cannot completely pre-
vent the domain gap between the source and target
domains. Therefore, we propose a method to miti-
gate the domain gap using quantum vectors from SQ-
VAE (Y.Takida et al., 2022). SQ-VAE is a method for
reconstructing high-resolution input images, capable
of representing images using only quantum vectors.
Figure 1 shows the overview of DG using SQ-
VAE. In Step 1, we divide N quantum vectors into
K groups, where N represents the number of quan-
tum vectors and K represents the number of cate-
gories. When an input feature is assigned to the quan-
tum vector defined as category 0, the feature is pre-
dicted as category 0. In Step 2, we use ground truth
labels to group the features into K categories and train
the model to bring these groups of the same category
closer together. Step 3 is inference. Since we do not
have access to the target domain, a domain gap arises.
However, by aligning the groups of each category, we
can minimize the domain gap. Even if there is a gap
in the unseen target domain, the features of the target
domain are likely to be assigned to the same or simi-
lar quantum vectors as those in the source domain and
thus categorized similarly. By using these methods,
we can prevent accuracy degradation due to unseen
target domains.
Experiments were conducted to segment blood
vessels from retinal image datasets (Drive (J.Staal
et al., 2004), Stare (A.Hoover et al., 2000),
Chase (G.Jiaqi et al., )). Each dataset has a differ-
ent domain due to varying imaging devices. Two
retinal image datasets were used for training, while
the remaining one was utilized for evaluation. The
proposed method achieved an average improvement
of 1.36% in mIoU compared to the original U-Net
when it served as the feature extractor, with a no-
table average increase of 2.71% in vascular regions.
When we use UCTransNet as a feature extractor, the
proposed method improved mIoU by 1.02% over the
original UCTransNet, with a significant improvement
of 2.17% in vascular regions.
Another experiment was conducted on
MoNuSeg (N.Kumar et al., 2020) dataset, which
exhibits diversity in nuclei across multiple organs
and patients and is captured under varying staining
methods at different hospitals. Therefore, DG is
required to extract features that are independent
of domain differences. Compared to the original
U-Net (O.Ronneberger et al., 2015), our method
using U-Net as a feature extractor improved mIoU by
2.53%, with an improvement of 2.73% in cell nuclei.
Additionally, compared to UCTransNet (H.Wang
et al., 2022), the proposed method using UCTransNet
as a feature extractor also improved mIoU by 3.0%,
with an improvement of 3.65% in cell nuclei.
The structure of this paper is as follows. Section 2
describes related works. Section 3 explains the details
of the proposed method. Section 4 presents and dis-
cusses the experimental results. Section 5 describes
conclusions and future work.
2 RELATED WORKS
2.1 Domain Generalization for
Semantic Segmentation
DG only allows access to the source domain and ex-
tracts features that are not specific to it (e.g., back-
ground or cell nuclei), mitigating domain shifts for
unseen target domains. Existing research on DG
for semantic segmentation often focuses on meth-
ods that remove domain-specific style information.
Techniques such as normalization (X.Pan et al.,
2018; S.Bahmani et al., 2022), whitening (S.Choi
et al., 2021), and diversification (Y.Zhao et al., 2022;
D.Peng et al., 2021) have been used to achieve DG.
However, WildNet argued that style information and
content information are not orthogonal, and whiten-
ing style information can inadvertently remove nec-
Domain Generalization Using Category Information Independent of Domain Differences
369
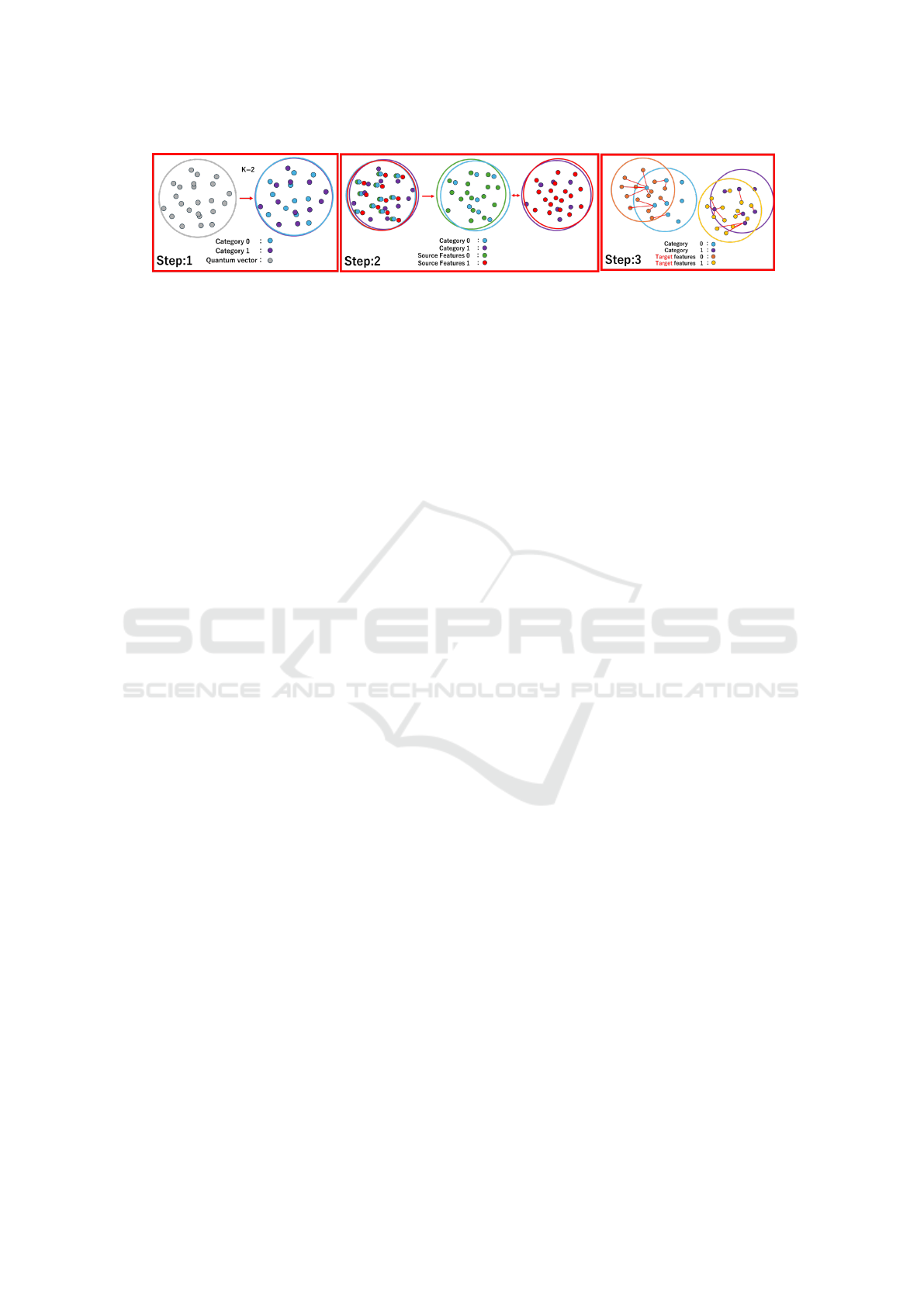
Figure 1: Overview of domain generalization using quantum vectors. This figure explains learning method for quantum
vectors used to absorb domain gaps and method to handle unseen target domains during inference.
essary content information. Therefore, to build a
high-precision model, it is essential to effectively ac-
quire information that is independent of the dataset.
We propose a method that splits the feature maps
from input images into two parts and trains them to
decorrelate from each other. One feature map is con-
strained to acquire category information independent
of domain differences, while the other retains source
domain-specific information. This approach allows
effective segmentation using feature maps that con-
tain category information independent of domain dif-
ferences.
2.2 Image Generation Model
Research on generative models has been extensive,
with various approaches proposed, such as Variational
Autoencoder (VAE) (Kingma and Welling, ), and
Vector Quantized VAE (VQ-VAE) (den Oord et al.,
2017). VAE maps input data to a latent space as a
probability distribution and samples latent variables
from this distribution to generate images. VQ-VAE
possesses higher quality image generation capabilities
and clustering abilities compared to VAE. However,
VQ-VAE has a non-differentiability issue due to the
use of the argmax function for discretization. This
problem was addressed by SQ-VAE. SQ-VAE uses
Gumbel-Softmax (E.Jang et al., 2016) to approximate
a categorical distribution in a differentiable manner,
allowing uninterrupted backpropagation. We propose
a method focusing on the clustering capability of SQ-
VAEs to bridge the domain gap between a source do-
main and an unseen target domain. Specifically, we
constrain the probabilities to divide the N quantum
vectors into K groups. Aligning the groups for each
category can minimize the gap with the unseen target
domain. This approach helps mitigate the accuracy
degradation caused by the target domain.
3 PROPOSED METHOD
When conventional semantic segmentation learns
from a specific dataset (source domain) and eval-
uates on an unseen dataset (target domain), accu-
racy decreases significantly due to domain differ-
ences such as different imaging devices and stain-
ing methods. To improve accuracy through DG, we
propose two approaches. First, we separate feature
maps into domain-independent category information
and domain-specific information of the source do-
main. Using domain-independent category informa-
tion for segmentation, we believe that accuracy is in-
dependent of the dataset domain used for training.
Second, using only category information cannot com-
pletely bridge the domain gap between the source and
target domains. Therefore, we propose a method to
address the domain gap using the quantum vectors of
SQ-VAE.
3.1 Category Information Independent
of Domain Differences
Figure 2 illustrates the overview of our method. To
extract information independent of domain differ-
ences, an input image x ∈ R
3×H×W
is processed by
a feature extractor, such as U-Net or UCTransNet,
which outputs the feature map Z ∈ R
C×H×W
. The
feature extractor, such as U-Net or UCTransNet,
achieves high accuracy and provides output images
of the same dimensions as the input images. This is
why we use U-Net or UCTransNet as the encoder.
The feature maps obtained from the encoder are di-
vided along the channel dimension into two parts: the
domain-independent category information and the re-
maining information. The divided feature maps are
denoted as Z
1
∈ R
C
1
×H×W
and Z
2
∈ R
C
2
×H×W
where
C
1
= C
2
= C/2. To separate domain-independent cat-
egory information from source domain-specific infor-
mation, the model is trained to decorrelate the feature
maps Z
1
and Z
2
. In this paper, we adopt DeepCCA,
which can learn nonlinear relationships, allowing it to
handle more complex data and achieve high precision
in removing correlations.
DeepCCA maximizes the correlation between two
variables in a nonlinear manner. However, since our
goal is to decorrelate them, squaring the output of
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
370

Figure 2: Overview of the proposed method. We extracted domain-independent category information to address unseen target
domains. Domain-independent category information is represented using Z
1
, which is used for segmentation. Additionally,
quantum vectors are used for quantization to bridge the gap between source domain and unseen target domain.
DeepCCA brings the result closer to zero.
L
corrcoe f
= corrcoe f (Z
1
,Z
2
)
2
(1)
After decorrelation, feature maps are quantized using
quantization vectors e
α
∈ R
N×C
1
and e
β
∈ R
N×C
2
. N
is the dimension of the embedding vector space. The
details are presented in Section 3.2. The reason for
preparing e
α
and e
β
is to assign them different roles.
e
α
is used to provide domain-independent categori-
cal features, while e
β
supplies source domain-specific
features. This approach allows each to serve distinct
functions. The feature maps after quantization are de-
noted as Z
′
1
and Z
′
2
. Z
′
1
and Z
′
2
are also trained to be
decorrelated using DeepCCA.
L
corrcoe f
= corrcoe f (Z
′
1
,Z
′
2
)
2
(2)
By using constraints in Equations 1 and 2, the
quantization vectors e
α
and e
β
become automatically
uncorrelated. By training Z
1
and Z
2
to be uncorre-
lated, they can assume different roles. For exam-
ple, if we assume that Z
1
and e
α
represent domain-
independent category features, then Z
2
and e
β
repre-
sent source domain-specific features. We hypothesize
that using these domain-independent features for seg-
mentation will improve accuracy. This method in-
volves training multiple features to be uncorrelated,
but there is no guarantee that Z
1
represents domain-
independent category information.
To address this, we propose a method to group fea-
tures within Z
1
that belong to the same category, em-
bedding domain-independent category information.
Specifically, we introduce a set of learnable represen-
tative vectors, t
K
∈ R
K×C
1
. Here, K denotes the num-
ber of categories. For example, for category t
0
, we
train the model to cluster features Z
0
1
in Z
1
with a tar-
get label of 0. This process is repeated for all K cat-
egories. Additionally, to ensure that these represen-
tative vectors do not capture source domain-specific
information, we train the model to separate the rep-
resentative vectors from each other. By doing so, we
can densely embed domain-independent category in-
formation.
L
domain
=
K−1
∑
i=0
n
||t
i
− Z
i
1
||
2
−
K−1
∑
j=0
||t
i
−t
j
||
2
2
o
(3)
where Z
0
1
and Z
1
1
are defined as the features of Z
1
when the target label is 0 and 1.
3.2 Segmentation Using SQ-VAE
In Section 3.1, we proposed a method that divides
features into those related to domain-independent
categories and those specific to the source domain.
Although only domain-independent features provide
DG capabilities, this does not completely prevent the
domain gap. Thus, we propose a method to bridge the
domain gap between the source and unseen target do-
mains using quantum vectors from the SQ-VAE. The
reason for using SQ-VAE is that it can generate vari-
ous clusters by utilizing quantum vectors. Addition-
ally, SQ-VAE is more accurate as an image genera-
tion model compared to VAE and VQ-VAE. We con-
strain the N quantum vectors from the SQ-VAE into
K groups to address the domain gap. For instance,
in the category of cell nuclei, we train the model to
bring the group of quantum vectors representing cell
nuclei closer to the group of features obtained from
input images of cell nuclei. By aligning these groups,
Domain Generalization Using Category Information Independent of Domain Differences
371

we can mitigate the domain gap between the source
and unseen target domains, allowing for better gener-
alization. We feed an image x ∈ R
3×H×W
into the fea-
ture extractor, such as U-Net and UCTransNet, which
outputs a feature map Z ∈ R
C×H×W
. As explained in
Section 3.1, to separate domain-independent category
features from domain-specific features, we divide the
feature map into two parts: Z
1
and Z
2
.
To address the domain gap, we define the quan-
tum vectors as e ∈ R
N×C/2
where C/2 is the chan-
nel dimension of the embedded vector e. We sepa-
rate domain-independent category information from
domain-specific in e
α
∈ R
N×C
1
and e
β
∈ R
N×C
2
. To
quantize the feature map obtained from the feature
extractor, we calculate the Mahalanobis distance be-
tween the feature map Z
1
and the quantum vector e
α
.
logit
1
= −
n
(e
α j
− Z
1
)
⊤
Σ
−1
γ
(e
α j
− Z
1
)
2
o
N−1
j=0
(4)
where Σ
γ
is a learnable parameter, and j refers to one
of the quantum vectors within the set of N vectors. It
is denoted as
∑
γ
= σ
2
γ
I. Then, logit
1
is expressed as
logit
1
=
||e
α j
− Z
1
||
2
2
2σ
2
γ
(5)
where logit
1
∈ R
HW ×N
is a matrix. In this case,
∑
γ
=
σ
2
γ
I is learned to approach zero from the initial value.
As training progresses, the probabilities of the dis-
tances between feature maps obtained by the encoder
and the quantum vectors become closer to a one-hot
encoding. This is similar to SQ-VAE. To probabilisti-
cally quantify the Mahalanobis distance between the
obtained feature map and the quantum vectors, we use
Gumbel-Softmax. We use Gumbel-Softmax because
it approximates the selection of discrete quantum vec-
tors as a continuous probability distribution and is dif-
ferentiable.
P
1
= Gumbelsoftmax
−
logit
1
τ
(6)
where τ is a learnable temperature parameter. Sim-
ilarly, we use Z
2
and e
β
to output P
2
. As shown in
Equations 5 and 6, we calculate the Mahalanobis dis-
tance and convert it to probabilities using Gumbel-
Softmax. These probabilities are then used to quan-
tize the features obtained from the encoder.
During evaluation, we replace the feature map ob-
tained from Equation 5 with the quantum vector that
has the closest Mahalanobis distance.
indices
1
= argmax(logit
1
) (7)
where argmax is used along the N-dimensional direc-
tion of logit
1
∈ R
HW ×N
.
The feature maps fed into the decoder are quan-
tized as Z
′
1
∈ R
C
1
×H×W
and Z
′
2
∈ R
C
2
×H×W
. Since the
channel dimension is split into two, the dimensions
from Z
′
1
and Z
′
2
are combined.
Z
′
= Concat(Z
′
1
,Z
′
2
) (8)
According to Equation 8, it becomes Z
′
∈ R
C×H×W
.
The decoder shown in Figure 2 uses an encoder-
decoder CNN (V.Badrinarayanan et al., 2017) with-
out skips. This is because U-Net or UCTransNet fea-
tures transmission mechanisms that allow the input
image to flow through easily, simplifying reconstruc-
tion and hindering the learning of intermediate layers.
The input Z
′
is passed through the encoder-decoder
CNN, producing the final output x
′
. The reconstruc-
tion error between the input image x and the final out-
put image x
′
is then calculated.
L
mse
=
∑
n
i=1
log(x
i
− x
′
i
)
2
2
(9)
where n is the total number of pixels in the input im-
age. The reason for adding log to the loss is that
the gradient becomes larger as x decreases. In other
words, we believe that the model should focus on finer
details during reconstruction. This is consistent with
the implementation of SQ-VAE.
The objective is to use features related to cate-
gories that are independent of domain differences and
employ quantum vectors to prevent domain gaps for
unseen target domains. Additionally, segmentation is
performed by the indices of the quantum vectors. This
is executed using Z
1
and e
α
∈ R
N×C
1
. It is divided
into K parts to separate roles based on the number of
segmentation categories K.
In this paper, we consider the case where K=2.
First, it is divided into two parts: e
0
α
∈ R
N
1
×C
1
, corre-
sponding to category label 0, and e
1
α
∈ R
N
2
×C
1
, cor-
responding to category label 1. Let N
1
and N
2
be
such that N
1
= N
2
= N/2. From Equation 6, we have
P
1
∈ R
H×W ×N
. When the category label is 0, we want
the probability of selecting e
0
α
to be 1. Similarly, when
the category label is 1, we want the probability of se-
lecting e
1
α
to be 1. To achieve this, we train the model
such that the sum of probabilities in the N
1
dimen-
sional direction of P
1
is 1 when the category label is
0, and the sum of probabilities in the N
2
dimensional
direction of P
1
is 1 when the category label is 1. In
other words, when the category label is 0, the model is
trained to minimize the distance between the feature
map Z
0
1
∈ R
C
1
×H
0
×W
0
related to the category label and
e
0
α
∈ R
N
1
×C
1
where H
0
and W
0
correspond to category
label 0. When the category label is 1, the model is
trained to minimize the distance between the feature
map Z
1
1
∈ R
C
1
×H
1
×W
1
related to the category label and
e
1
β
∈ R
N
2
×C
1
where H
1
and W
1
correspond to category
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
372

Figure 3: Weights that are learned to focus on parts where
predictions become uncertain. Treat red as weight.
label 1. This learning method is as
L
code
= log
n
w
0
× (1 −
N
2
−1
∑
N=0
P
1
)
2
L=0
+w
1
× (1 −
N−1
∑
N=
N
2
P
1
)
2
L=1
o
(10)
where L is the label, and w
0
and w
1
are weights added
to prioritize uncertain predictions. By using these
weights, the model can focus on learning parts where
predictions are uncertain, such as the boundaries in
segmentation.
Figure 3 shows a conceptual diagram of the
weights. The maximum values of the prediction prob-
abilities from index 0 to (N-1)/2 and from N/2 to N-
1 are obtained. These are defined as max(P
0
1
) and
max(P
1
1
), respectively. Next, the absolute value of the
difference between these maximum values is taken as
di f = |max(P
0
1
) − max(P
1
1
)| (11)
The absolute differences are collected for all pixels
and divided by the maximum value among all pixels
to normalize them from 0 to 1. The weight of the i-th
pixel is as follows:
w
i
=
di f
i
max(di f )
(12)
The weight w is less likely to be learned if it is closer
to 1, as it indicates higher certainty. Conversely, if it is
closer to 0, it signifies greater uncertainty in the pre-
diction, and it is learned more intensively. The reason
weights closer to 0 are learned more intensively than
those closer to 1 is that Equation 10 includes a log,
making smaller loss values more significant. Taking
the derivative of log(x) results in 1/x, meaning that
as x becomes smaller, the gradient becomes larger,
thereby making smaller loss more significant.
Finally, the index of the quantization vector clos-
est to the feature vector is obtained, which is related
to the category and independent of the domain dif-
ferences. If this index is between 0 and (N/2) − 1,
category label 0 is assigned. If the index is between
N/2 and N −1, category label 1 is assigned. This cat-
egory label is used as the final segmentation predic-
tion. The learning method using this quantized vector
is shown in Figure 1, with Step 2 particularly pertain-
ing to that part. When the category label is 0, the
model is trained so that the sum in the N
1
dimensional
direction of P
1
equals 1. When the category label is 1,
the model is trained so that the sum in the N
2
dimen-
sional direction of P
1
equals 1. This approach divides
the features into two groups using the labeled data,
similar to dividing the quantization vectors into two
groups. The model is then trained to bring the groups
closer together. As a result, even in the presence of a
domain gap in the unseen target domain, it can be mit-
igated, as quantization assigns similar quantum vec-
tors to clusters (groups of the same category).
4 EXPERIMENTS
4.1 Implementation Details
Our method is evaluated on blood vessel segmenta-
tion from three types of fundus images: Drive, Stare,
and Chase. The Drive, Stare, and Chase datasets each
contain a total of 20, 20, and 28 images, respectively,
along with annotations for segmenting the images into
classes: background and blood vessels. The images
in the Drive dataset were captured using a Canon CR5
non-mydriatic 3CCD camera. The images in the Stare
dataset were obtained using a Top Con TRV-50 reti-
nal camera. The images in the Chase dataset were
captured with a Nidek NM-200-D fundus camera. As
these images were taken with different cameras, they
can be considered to belong to different domains.
Of the three datasets, two are used for training,
while the remaining dataset is used for evaluation. By
rotating this arrangement, the DG performance is as-
sessed. Each dataset is divided into five parts to per-
form 5-fold cross-validation, with four parts used for
training (e.g., 4/5 of Drive and 4/5 of Chase) and one
part used for validation (e.g., 1/5 of Drive and 1/5 of
Chase). Subsequently, the two datasets used for train-
ing and validation are combined to ensure there is no
data imbalance. The validation data from the remain-
ing dataset, which was not used for training, is used
for evaluation (e.g., 1/5 of Stare). However, since the
Chase dataset contains more images, to avoid bias in
training or evaluation, the number of images in the
Chase dataset is randomly reduced to 20 for the ex-
periments.
Additionally, we conduct experiments on the
MoNuSeg dataset, which contains tissue images of
tumors from various organs diagnosed in several pa-
tients across multiple hospitals. Due to the diverse
appearance of nuclei across different organs and pa-
tients, as well as the variety of staining methods
used by various hospitals, it is important to extract
domain-agnostic information from this dataset. The
Domain Generalization Using Category Information Independent of Domain Differences
373

MoNuSeg dataset consists of 30 images for training
and 14 images for evaluation. Among the training
data, 24 images are allocated for training, and 6 im-
ages are reserved for validation. The test data is used
as is for evaluation and includes lung and brain cells
that are not present in the training data, rendering
them unseen data.
For all experiments, the seed value is changed four
times to calculate average accuracy. We resize all im-
ages to 256 × 256 pixels as preprocessing. The learn-
ing rate is set to 1 × 10
−3
, the batch size is 2, the op-
timizer is Adam, and the number of epochs is 200.
We used an Nvidia RTX A6000 GPU. The number of
quantum vectors is set to 512. The evaluation metric
is intersection over union (IoU), and we evaluate us-
ing the IoU for each class and the mean IoU (mIoU)
across all classes. We compared the proposed method
with U-Net and UCTransNet. The rationale is that the
proposed method uses U-Net or UCTransNet as an
encoder and makes predictions using quantum vectors
based on its output. In other words, the same feature
extractor is used up to the point of segmentation pre-
diction.
4.2 Domain Generalization on Chase,
Stare, and Drive Datasets
The results of DG on the Chase, Stare, and Drive
datasets are shown in Table 1. The method with
the highest accuracy is shown in orange, while the
second-highest accuracy is in blue. When the Drive
and Stare datasets were used for training and the
Chase dataset for evaluation, the proposed method
(U-Net+ours) using U-Net as a feature extractor ex-
hibited a 1.80% improvement in mIoU compared
to the original U-Net, with a specific improvement
of 3.69% in the blood vessel area. Additionally,
the proposed method (UCTransNet+ours) using UC-
TransNet as a feature extractor demonstrated a 2.41%
improvement in mIoU compared to the original UC-
TransNet, with a specific improvement of 5.01% in
the blood vessel area. When the Drive and Chase
datasets were used for training and the Stare dataset
for evaluation, the proposed method (U-Net+ours) us-
ing U-Net as a feature extractor showed a 1.20% im-
provement in mIoU compared to the original U-Net,
with a specific improvement of 2.47% in the blood
vessel area. Additionally, the proposed method (UC-
TransNet+ours) using UCTransNet as a feature ex-
tractor noted a 0.32% improvement in mIoU com-
pared to the original UCTransNet, with a specific im-
provement of 0.76% in the blood vessel area. When
the Stare and Chase datasets were used for training
and the Drive dataset for evaluation, the proposed
Table 1: IoU and standard deviation Chase, Stare, and Drive
datasets. orange indicates the highest accuracy, and blue
indicates the second-highest accuracy.
datasets methods background blood vessels mIoU
Chase
U-Net 95.33(±0.35) 43.56(±4.14) 69.44(±2.15)
U-Net + ours 95.24(±0.48) 47.25(±1.82) 71.24(±1.13)
UCTransNet 95.27(±0.37) 45.92(±3.84) 70.59(±1.97)
UCTransNet + ours 95.06(±0.39) 50.93(±1.65) 73.0(±0.92)
Stare
U-Net 95.81(±0.86) 56.70(±6.79) 76.26(±3.73)
U-Net + ours 95.75(±0.71) 59.17(±4.16) 77.46(±2.36)
UCTransNet 95.80(±0.85) 56.71(±5.66) 76.25(±3.20)
UCTransNet + ours 95.67(±0.71) 57.47(±4.17) 76.57(±2.36)
Drive
U-Net 95.56(±0.53) 57.86(±1.85) 76.71(±1.17)
U-Net + ours 95.75(±0.40) 59.84(±3.06) 77.80(±1.57)
UCTransNet 95.62(±0.49) 58.35(±2.10) 76.99(±1.28)
UCTransNet + ours 95.58(±0.46) 59.10(±1.58) 77.34(±1.01)
method (U-Net+ours) using U-Net as a feature ex-
tractor showed a 1.09% improvement in mIoU com-
pared to the original U-Net, with a specific improve-
ment of 1.98% in the blood vessel area. Addition-
ally, the proposed method (UCTransNet+ours), using
UCTransNet as a feature extractor showed a 0.35%
improvement in mIoU compared to the original UC-
TransNet, with a specific improvement of 0.75% in
the blood vessel area. These improvements indicate
that DG is effectively achieved.
Additionally, segmentation results are shown in
Figure 4. The top three rows display the results on
the Chase, Stare, and Drive datasets. The areas high-
lighted in red boxes show significant improvements.
For the Chase dataset, vascular regions in the red
box of the input image appear slightly darker, which
the original U-Net and UCTransNet predict as back-
ground. In contrast, our methods (U-Net+ours and
UCTransNet+ours) extract category information in-
dependently of the domain, preventing domain gaps,
can densely extract blood vessel category informa-
tion, predicting them correctly. For the Stare dataset,
focusing on the red box areas, the original U-Net
and UCTransNet make predictions indicating discon-
nected blood vessels. However, the proposed meth-
ods (U-Net+ours and UCTransNet+ours) predict con-
nected blood vessels, effectively extracting category
information independently of the domain. For the
Drive dataset, in the red box areas, the original U-
Net and UCTransNet predict the blood vessels as thin
or disconnected. In contrast, the proposed methods
predict thicker blood vessels and connect previously
disconnected vessels, successfully extracting domain-
independent information.
4.3 Domain Generalization on
MoNuSeg
The results of DG on the MoNuSeg dataset are shown
in Table 2. The method with the highest accuracy
is highlighted in orange, while the method with the
second-highest accuracy is shown in blue. Compar-
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
374

Figure 4: Segmentation results on Chase, Stare, MoNuSeg, and Drive datasets. From left to right, the images show input
images, ground truth, results by original U-Net, our method (U-Net+ours), original UCTransNet, and our method (UC-
TransNet+ours).
Table 2: IoU and standard deviation on MoNuSeg dataset.
orange method achieved the highest accuracy, while blue
method attained the second-highest accuracy.
methods background cell nucleus mIoU
U-Net 87.36(±0.96) 61.55(±1.17) 74.46(±1.01)
U-Net + ours 89.71(±1.24) 64.28(±1.10) 76.99(±1.17)
UCTransNet 87.79(±0.07) 60.93(±0.51) 74.36(±0.23)
UCTransNet + ours 90.15(±0.95) 64.58(±0.67) 77.36(±0.80)
ison results between conventional methods and the
proposed methods (methods + ours) are included. As
a result, the proposed method (U-Net+ours) achieved
a 2.53% improvement in mIoU compared to the orig-
inal U-Net, with a specific improvement of 2.73%
in the cell nucleus area. Additionally, the pro-
posed method (UCTransNet+ours) achieved a 3.0%
improvement in mIoU compared to the original UC-
TransNet, with a specific improvement of 3.65% in
the cell nucleus area. These results demonstrate
strong generalization performance to unseen target
domains. The improved accuracy in cell nuclei, in
particular, suggests that the method can effectively
handle the diversity in the appearance of cell nuclei
and staining methods across different domains, suc-
cessfully extracting cell nucleus-specific features.
Segmentation results from conventional methods
and our methods are displayed in the bottom row of
Figure 4. The areas highlighted in red boxes indicate
where significant improvements were observed. The
original U-Net struggled with DG, often predicting
background as cell nuclei. In contrast, the proposed
method (U-Net+ours) successfully extracted category
information independent of the domain, closing do-
main gaps and enabling accurate predictions. Simi-
larly, the original UCTransNet over-predicted cell nu-
clei in the areas highlighted in red. However, the
proposed method (UCTransNet+ours) effectively ex-
tracted information on various cell nuclei, leading to
accurate predictions.
5 CONCLUSION
We proposed a method that generalizes well to
datasets with differences in imaging equipment and
staining methods (target domain) compared to the
dataset (source domain) on which the model was
trained. The proposed method showed significant im-
provements on cell image datasets with various stain-
ing methods and fundus images captured by different
imaging devices. These results demonstrate the gen-
eralization performance of our method to unseen tar-
get domains. In the future, we would like to evaluate
our method on a multi-class segmentation problem.
ACKNOWLEDGMENTS
This paper is partially supported by the Strategic In-
novation Creation Program.
Domain Generalization Using Category Information Independent of Domain Differences
375

REFERENCES
A.Hoover et al. (2000). Locating blood vessels in retinal
images by piece-wise threshold probing of a matched
filter response. IEEE Transactions on Medical Imag-
ing, 19(3):203–210.
B.Cheng et al. (2022). Masked-attention mask transformer
for universal image segmentation. In Proceedings
of the IEEE/CVF conference on computer vision and
pattern recognition, pages 1290–1299.
den Oord, A. et al. (2017). Neural discrete representation
learning. In I.Guyon et al., editors, Advances in Neu-
ral Information Processing Systems, volume 30. Cur-
ran Associates, Inc.
D.Peng et al. (2021). Global and local texture random-
ization for synthetic-to-real semantic segmentation.
IEEE Transactions on Image Processing, 30:6594–
6608.
E.Jang et al. (2016). Categorical reparameterization with
gumbel-softmax. arXiv preprint arXiv:1611.01144.
F.Milletari et al. (2016). V-net: Fully convolutional neural
networks for volumetric medical image segmentation.
In 2016 fourth international conference on 3D vision
(3DV), pages 565–571. Ieee.
Furukawa, R. and Hotta, K. (2021). Localized feature ag-
gregation module for semantic segmentation. In 2021
IEEE International Conference on Systems, Man, and
Cybernetics (SMC), pages 1745–1750. IEEE.
G.Andrew et al. (2013). Deep canonical correlation analy-
sis. In Dasgupta, S. and McAllester, D., editors, Pro-
ceedings of the 30th International Conference on Ma-
chine Learning, volume 28 of Proceedings of Machine
Learning Research, pages 1247–1255, Atlanta, Geor-
gia, USA. PMLR.
G.Jiaqi et al. Chase: A large-scale and pragmatic chinese
dataset for cross-database context-dependent text-to-
sql.
H.Wang et al. (2022). Uctransnet: rethinking the skip
connections in u-net from a channel-wise perspective
with transformer. In Proceedings of the AAAI con-
ference on artificial intelligence, volume 36, pages
2441–2449.
J.Staal et al. (2004). Ridge-based vessel segmentation in
color images of the retina. IEEE transactions on med-
ical imaging, 23(4):501–509.
J.Wang et al. (2020). Deep high-resolution representa-
tion learning for visual recognition. IEEE transac-
tions on pattern analysis and machine intelligence,
43(10):3349–3364.
Kingma, D. P. and Welling, M. Auto-encoding variational
bayes.
N.Kumar et al. (2020). A multi-organ nucleus segmentation
challenge. IEEE Transactions on Medical Imaging,
39(5):1380–1391.
O.Ronneberger et al. (2015). U-net: Convolutional
networks for biomedical image segmentation. In
Medical Image Computing and Computer-Assisted
Intervention–MICCAI 2015: 18th International Con-
ference, Munich, Germany, October 5-9, 2015, Pro-
ceedings, Part III 18, pages 234–241. Springer.
P.Wang et al. (2023). One-peace: Exploring one gen-
eral representation model toward unlimited modali-
ties. arXiv preprint arXiv:2305.11172.
S.Bahmani et al. (2022). Semantic self-adaptation: Enhanc-
ing generalization with a single sample. arXiv preprint
arXiv:2208.05788.
S.Choi et al. (2021). Robustnet: Improving domain gen-
eralization in urban-scene segmentation via instance
selective whitening. In Proceedings of the IEEE/CVF
conference on computer vision and pattern recogni-
tion, pages 11580–11590.
Shibuya, E. and Hotta, K. (2020). Feedback u-net for cell
image segmentation. In Proceedings of the IEEE/CVF
Conference on computer vision and pattern recogni-
tion workshops, pages 974–975.
S.Lee et al. (2022). Wildnet: Learning domain generalized
semantic segmentation from the wild. In Proceedings
of the IEEE/CVF conference on computer vision and
pattern recognition, pages 9936–9946.
V.Badrinarayanan et al. (2017). Segnet: A deep convo-
lutional encoder-decoder architecture for image seg-
mentation. IEEE transactions on pattern analysis and
machine intelligence, 39(12):2481–2495.
X.Pan et al. (2018). Two at once: Enhancing learning
and generalization capacities via ibn-net. In Proceed-
ings of the european conference on computer vision
(ECCV), pages 464–479.
Y.Liu et al. (2020). Efficient semantic video segmentation
with per-frame inference. In Computer Vision–ECCV
2020: 16th European Conference, Glasgow, UK, Au-
gust 23–28, 2020, Proceedings, Part X 16, pages 352–
368. Springer.
Y.Takida et al. (2022). SQ-VAE: Variational Bayes on
discrete representation with self-annealed stochastic
quantization. In Chaudhuri, K., Jegelka, S., Song,
L., Szepesvari, C., Niu, G., and Sabato, S., edi-
tors, Proceedings of the 39th International Conference
on Machine Learning, volume 162 of Proceedings
of Machine Learning Research, pages 20987–21012.
PMLR.
Y.Zhao et al. (2022). Style-hallucinated dual consistency
learning for domain generalized semantic segmenta-
tion. In European conference on computer vision,
pages 535–552. Springer.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
376
