
Novel Approach to De-Identify Relational Healthcare Databases at Rest:
A De-Identification of Key Data Approach
Yazeed Ayasra
a
, Mohammad Ababneh
b
and Hazem Qattous
c
Princess Sumaya University for Technology, Amman, Jordan
yazeed.ayasra@gmail.com, {m.ababneh, h.qattous}@psut.edu.jo
Keywords:
Health Information Security, Relational Database Security, Data Anonymization, ePHI Security, HIPAA
Compliance.
Abstract:
Health information systems are widely used in the healthcare sector, and migration to cloud-based applica-
tions continues to be prominent in recent practices. Various legislations were issued by different countries to
ensure the confidentiality of personal health information, introducing liabilities, and imposing penalties and
fines to organizations in violation. This drives organizations to deploy significant investments in information
security to safeguard various health information systems. The healthcare industry has experienced the second
highest data breaches compared to other industries at 24.5% of the total data breaches in the United States
between 2005 and 2023. Database layer vulnerabilities remain one of the most exploited resulting in attacks
causing devastating confidentiality breaches for electronic personal health information (ePHI). The framework
suggested in this work relied on de-identification using the health insurance portability and accountability act
(HIPAA) safe harbor method of removing 18 identifying attributes from the data in its resting state. To achieve
this, the work proposes 7 rules that allow the migration of health information system databases to the suggested
framework structure to maintain a de-identified state of the database at rest. This is achieved through the seg-
regation of identifying information in different tables based on their identification power and frequency of
use while structuring them in a hierarchical manner where tables refer to the next or previous levels through
encrypted foreign keys. The paper extends to successfully transform a typical EHR system database schema
into a de-identified version of itself abiding to the 7 rules suggested by this work.
1 INTRODUCTION
This To tackle the various issues of ePHI security,
the US Congress acted and passed the Health Insur-
ance Portability and Accountability Act (HIPAA) in
1996. The main objective of HIPAA was to estab-
lish a uniform set of standards for the disclosure of
health data by healthcare providers. The act also sets
standards for various other aspects including infor-
mation security, privacy, fraud prevention, electronic
data exchange, healthcare access, revenue, and in-
surance (Miller and Payne, 2016). HIPAA imposes
penalties and fines on organizations in breach of its
security and privacy rules reaching up to 1.5M USD
per incident. This drives organizations to deploy sig-
nificant investments in information security to safe-
guard various health information systems. One way
to perform data sharing while abiding by HIPAA reg-
a
https://orcid.org/0000-0001-9283-7251
b
https://orcid.org/0000-0002-6431-2201
c
https://orcid.org/0000-0002-8468-1602
ulations is the production of de-identified datasets.
De-identified datasets are defined as health informa-
tion that does not have any reasonable basis to believe
that identification of individuals using it is possible
(Caplan, 2003). HIPAA requires validation of de-
identified datasets through one of two methods: ex-
pert determination or safe harbor. Expert determina-
tion requires a formal inspection and approval by a
qualified subject matter expert, while the safe harbor
method requires the removal of 18 personal health in-
formation identifiers. Limited datasets are also widely
used for research purposes. Limited datasets are lim-
ited sets of identifiable patient information that are
considered personal health information because of the
possibility of re-identification as per the HIPAA pri-
vacy rule (Caplan, 2003). Most recent technologies
in cloud applications relied on the three-tier approach
with an SQL Server as the database server. Three-
tier setups improve performance and limit the num-
ber of attacks on databases due to their segregation
from the application layer (Singh, 2024). Although
Ayasra, Y., Ababneh, M. and Qattous, H.
Novel Approach to De-Identify Relational Healthcare Databases at Rest: A De-Identification of Key Data Approach.
DOI: 10.5220/0013302000003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 207-218
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
207

this architecture provides an extra layer of protection
for database servers, it is still prone and vulnerable
to all common database server attacks through the
application layer and directly on the database layer
(OWASP, 2024). Piggy-backed queries - where an at-
tacker attempts to piggyback additional statements to
the pre-existing SQL queries – are very common and
constitute most SQL attacks (Sai Lekshmi and De-
vipriya, 2017), SQL injections constitute 15% of at-
tacks against cloud-based applications (Unlu and Bi-
cakci, 2010). Such attacks and others directly con-
tribute to risks against information integrity, avail-
ability, and confidentiality. In the healthcare context
data leaks result in breaches of confidentiality and pri-
vacy of electronic personal health information expos-
ing patients and organizations to significant liabili-
ties. Newer DBMS versions support a variety of fea-
tures to prevent common attacks such as stored proce-
dures that can help prevent SQL injection attacks but
only when stored procedures are implemented cor-
rectly (Patel et al., 2020). Although such features
would increase database security, vulnerabilities are
still present and database information is still acces-
sible to internal resources managing them. In the
United States, 35,167 known data breaches have been
reported from 2005 to 2023 (Rights, 2023).
Figure 1: Proposed methodology by (Rights, 2023).
The healthcare industry has experienced the sec-
ond most data breaches compared to other industries
at 24.5% of the total data breaches (Rights, 2023).
From 2020 to 2022, the industry suffered from 1,046
data breaches in the United States alone, costing an
average of $6.45 million per incident (Enterprise,
2018).
Anonymization and de-identification of data is
one solution adopted by the healthcare community
to produce datasets for use in the research communi-
ties. De-identified datasets enable information shar-
ing for research and public health purposes but can-
not be used to protect data at rest for various health
information systems. This is due to the integral need
for personal health information identification for con-
tinuity of care purposes. This work explores a method
to de-identify data at rest and allow re-identification at
the application layer.
Various security methods, models and new tech-
nologies are widely used to protect applications and
databases from unauthorized access or data leaks.
Mandatory Access Control (MAC) is an access con-
trol model enabling access to information based on
confidentiality of information and clearance level.
Encryption converts information to ciphertext al-
lowing access to authorized users and applications
in custody of the key to decipher the code and
access the original information. Blockchain is a
distributed database composed of blocks accessed
through the acquisition of cryptographic hashes to re-
veal stored information. The framework proposed in
this work attempts to utilize MAC, encryption and
a blockchain-like structure to de-identifty electronic
personal health information in relational databases.
The usage of cloud-based platforms has expo-
nentially increased in the past few years due to the
increasing need for medical data available online
(Chhabra et al., 2022). Various types of cloud-based
platforms were required for a variety of clinical and
medical applications such as contact tracing, labora-
tory test results, travel health declarations, clinical en-
counters, and other public health tools. This resulted
in a significant increase in the amount of electroni-
cally identifiable personal health information (ePHI)
available online that could be vulnerable to various
cloud-based application attacks. Such high availabil-
ity of health information exposes organizations to sig-
nificant implications and liabilities due to confiden-
tiality breaches of electronic personal health informa-
tion (ePHI) resulting in violations of health informa-
tion privacy policies and laws in various countries.
Four out of the top five vulnerabilities published
by the Open-Source Foundation for Application Se-
curity (OWASP) can directly impact the database
layer and cause unauthorized access to stored infor-
mation revealing confidential information. Broken
access control, injection, insecure design, and secu-
rity misconfigurations can all lead to privacy and con-
fidentiality breaches with devastating impacts on the
healthcare sector. Best practices such as parameter-
ized queries, stored procedures, and other solutions
are still vulnerable to a variety of attacks in certain
setups (OWASP, 2024).
The emerging needs of big data had rendered non-
relational (NoSQL) databases a widely used solution
for data storage and retrieval. However, relational
(SQL) databases remain the most widely used solu-
tion for data storage and retrieval in various applica-
tions (Capris et al., 2022). Those databases remain
prone to various attacks resulting in confidentiality
breaches for electronic personal health information in
HEALTHINF 2025 - 18th International Conference on Health Informatics
208

the healthcare sector.
SQL injections, physical attacks, access control
breaches, and other database server attacks could the-
oretically have less impact if released information is
de-identified. Achieving a de-identified storage sta-
tus using the safe harbor method by eliminating ac-
cess to the 18 known identifiers of electronic health
records in databases at rest would prevent policy vi-
olations. This would lessen liabilities for healthcare
organizations and help reduce patients’ confidential-
ity breaches while minimally impacting application
performance. Such a storage framework is also im-
mune to insider attacks and would offer flexibility in
producing limited and de-identified datasets
2 LITERATURE REVIEW
This section explores various modalities designed to
increase the immunity of relational databases against
common relational database attacks. It covers a vari-
ety of solutions that attempt to solve the problem from
different angles.
To produce de-identified and limited datasets,
(Erdal et al., 2012) discusses an original framework
that uses structured data in relational databases-based
health information systems. The process performs ex-
tract, transform, and load (ETL) to produce a limited
dataset that can later be processed into a de-identified
dataset.
The objective of this study is to produce datasets
that can be used for research purposes that are com-
pliant with HIPAA and the local IRB. The researchers
took an interesting approach that relied on the manual
removal of HIPAA-protected data attributes through a
variety of methods. To isolate patient identifiers, the
researchers produced a multi tiered mapping between
patient identifiers and pseudo identifiers with a layer
of one-way hashing that prevents re-identification
through classical SQL queries. For patients more than
90 years of age, the researchers stored their age in
a separate table while recording a maximum age of
89 at the level of the limited and de-identified. To
produce de-identified datasets, the researchers added
a random time increment to disguise dates. The pa-
per later proves the viability of their de-identification
framework by running the framework on 20 mil-
lion records ensuring the efficiency of the framework.
The comparison between the original dataset and the
produced dataset highlighted a significant increase
in CPU processing time (averaging 15 seconds per
execution) and a better performance when generat-
ing limited datasets in comparison with de-identified
datasets. This framework appears to be viable to re-
place the common manual de-identification processes
required to produce limited and de-identified datasets
for reporting and research purposes. However, such
an approach does not offer any layer of protection to
the original datasets, which are still at risk of common
relational database vulnerabilities. Furthermore, the
model does not offer a real-time reflection of the data
available as it would require a significant overhead if
deployed in a real-time production environment. The
proposed framework in this work offers a solution that
would minimally tax the application when protecting
health information systems databases.
The work presented in (Lin et al., 2016) at-
tempted to reach k-anonymity in relational databases
through an efficient algorithm. To implement this,
records go through an anonymization algorithm that
would transform data points into a more generic ver-
sion of themselves to resemble (k) number of rows.
The authors suggested an original framework that
performed anonymization through three consecutive
modules. The pre-processing module was designed
to encode transactions to bitmaps that are sorted us-
ing the Gray order to approximate similar and clas-
sify them into several segments based on similarity
for efficient anonymization. The Travelling Salesman
Problem (TSP) module was designed to find a cycli-
cal loop between various transactions in each segment
rendering the TSP module more efficient to find the
cyclical loop between a lesser number of transactions
at a time. The anonymization module achieves k-
anonymity through the replacement of each class with
its center class as calculated by the module. This
results in data loss, which is necessary to achieve
anonymization.
The paper extended to undergo an experimental
evaluation resulting in anonymization for five real-
world datasets and one synthetic dataset. Their re-
sults showed a significant increase in data loss with
an increased (k) value which is consistent with k-
anonymization by nature. Although the researchers
are presenting a reasonable algorithm that can achieve
anonymization, they illustrated significant data loss as
anticipated with such an approach. Such data loss can
be intolerable in electronic health records or medical
records systems.
Authors of (El Emam, 2010) suggested a method
to measure the re-identifiability of health information
based on a conceptual five-level model of the identifi-
ability continuum as shown in Figure 2. They concep-
tualize that datasets can reside in gray zones between
identifiable and non-identifiable statuses. The authors
classified health information datasets into five con-
ceptual categories that are ordinally classified based
on the possibility of re-identification. Level 1, which
Novel Approach to De-Identify Relational Healthcare Databases at Rest: A De-Identification of Key Data Approach
209

presents datasets that are inclusive of readily identi-
fiable data such as names, social security numbers,
and addresses, is at the base of their model with a
higher risk of re-identification. Level 2 constitutes
datasets with reversibly and irreversibly masked iden-
tifiers but accessible quasi-identifiers (i.e. dates, zip
codes, date of birth). Exposed data, or level 3 as
described by the suggested continuum, is masked
data that went through an obfuscation process for
quasi-identifiers rendering the dataset more resistant
to re-identification but still considered as a high-risk
dataset. This is because the custodian did not assess
re-identifiability at this level. Level 4 is considered by
the authors as the first level to host true de-identified
data. Aggregate extracts of the data were marked as
level 5, which is the highest form of de-identified data
as per the continuum. The paper argues that measur-
ing health data identifiability is possible based on de-
termining the possible risk associated with the data re-
quester. They extended to classify risks into three cat-
egories. Prosecutor risk is relevant when an adversary
(family member or knowledgeable personnel) is at-
tempting to re-identify data based on previous knowl-
edge of the subject and the fact that the subject exists
in the dataset. Journalist risk is relevant when an ad-
versary is attempting to re-identify based on the mere
existence of subject knowledge but not its existence
in the dataset. Finally, marketer risk, which repre-
sents attempts of mass re-identification when compar-
ing datasets with each other such as a comparison be-
tween a de-identified health data with a national reg-
istry of citizens, which could result in re-identification
of a subset of subjects in the dataset. The article ar-
gues that by establishing a custodian understanding of
the type of risk associated with each type of recipient
of the datasets, they would be capable of determining
a threshold on the identifiability continuum of types
of data at risk of re-identification.
Although the authors’ opinion of the existence of
re-identification of a dataset on a continuum is valid,
the suggestion that the risks are closely associated
with target recipients of data is not conclusive. Health
information is considered sensitive data with a signif-
icant attacker appetite. Therefore, it is prone to re-
siding with an audience that could be of interest in
exploiting information when the dataset is not prop-
erly de-identified. The framework proposed in this
work is designed to enable back-end applications to
produce all levels of datasets on the continuum with
the capacity to aggregate information into producing
level 5 datasets.
The authors of (Omran et al., 2009) discussed the
importance of health information exchange and the
various recent methods adopted to allow it among var-
Figure 2: Identifiability Continuum (El Emam, 2010).
ious health service providers. The paper also high-
lights that the higher the availability of healthcare
data, the more prone it is to confidentiality and in-
tegrity breaches. The Healthcare sector is benefiting
from the utilization of web services to promote in-
teroperability, but it significantly increases the possi-
bility of different attacks that would compromise the
confidentiality and privacy of patients.
The paper discussed security mechanisms and is-
sues in both databases and web services. The first
database security measure discussed was access con-
trol. The paper compared content-based access con-
trol that would only allow access to pre-tabulated
views, RBAC model, and System R access control.
They later proceed to discuss k-anonymity, aiming
to abstract data and release a version that is de-
identified. All subjects and attributes that can result in
the identification are omitted, leaving a de-identified
version of the data that could be shared with minimal
confidentiality and privacy risks.
Although there have been successful attempts
to re-identify patients from de-identified data, other
measures could be taken by policymakers and
database architects to minimize such inference. An
example of that is changing quantifiable attributes
such as age to nominal and ordinal data such as an
age group. K-anonymity solves the problem of health
information exchange to a great extent, but it does
not address security in the case of database vulner-
abilities of healthcare cloud applications. It results
in significant data loss. The authors also discussed
database encryption and Hippocratic databases, both
approaches increase the level of security with sig-
nificant performance impact in database encryption
methods.
The study continued to explain web services secu-
rity and suggested that network security should also
be addressed. As the scope of this work focuses on
dataset security in relational databases against suc-
cessful attacks, our focus was to study the different
database protection approaches in comparison to the
model proposed in this work.
Authors of (Omran et al., 2009) underwent a
case study of openEHR security and architecture.
openEHR is a widely used open-source electronic
health records system. The paper discusses some of
the security features of the application such as the
HEALTHINF 2025 - 18th International Conference on Health Informatics
210

segregation of demographics from clinical and ad-
ministrative data, version control, and access control
among others. The openEHR approach to the seg-
regation of data is to achieve anonymity by elimi-
nating any direct clues to identify the patients in the
EHR database. The application enforces the storage
of an EHR ID (ehr id) that serves as the foreign key
between two databases on different servers. This is
stored in a cross-reference database that could be en-
crypted. This approach attempts to achieve a simi-
lar goal to this work. However, the approach pre-
sented in this work offers a more efficient and cost-
effective solution since the storage of information is
done on one database server. It also allows differ-
ent levels of protection based on the sensitivity of the
various data attributes. Due to the hierarchical na-
ture of the proposed framework, software adapting
this framework would be capable of producing lim-
ited or de-identified datasets on demand without sig-
nificant overhead. The suggested framework enables
security policies and role-based access control due to
the multi-tiered classification of sensitive data.
The work presented in (Avireddy et al., 2012) dis-
cusses the utilization of randomized encryption for
the protection of SQL injection attacks. The paper
presented the risks of SQL injection attacks and their
common incidence for cloud applications. The pa-
per aimed to develop an algorithm that uses random-
ized encryption to minimize confidentiality breach
risk caused by SQL injection attacks. It extended to
develop an application that adapts the algorithm to
validate the immunity of this algorithm towards SQL
injection attacks.
The authors focused on SQL injection attacks
caused by poor validation on the application tier in
a three-tier cloud application setup. Such improper
validation might cause SQL statements to be passed
toward the database server, causing potential extrac-
tion of confidential data, taking control, or disturbing
data integrity. SQL injection attacks using tautology
are very common in applications that do not enforce
proper front-end and back-end validation. One ex-
ample of that would be an attacker passing an SQL
statement to extract data or alter the original query
designed by development teams. Another example of
such attacks is the usage of incorrect queries to return
errors from the database server. Attackers usually use
this method to infer information about the database
structure which could later be used during the con-
struction of malicious SQL statements.
The Random4 algorithm is what the authors pro-
pose as their contribution to this paper. It constitutes
a method that is based on randomization to convert
input into an encrypted salt. The algorithm chooses
specific characters that could be used in salting any
front-end input before passing it back to the database
engine. Each character in the input is later mapped
into one of 4 probable characters based on the pre-
defined lookup table relying on the character mapping
technique they chose to implement.
The paper suggests that encrypted input should be
stored within the database which would significantly
decrease the impact of SQL injection attacks. They
attempted to develop an application written in C# to
encrypt incoming inputs to provide examples. The
proposed model would prevent piggy-backed queries
since passed input is changed and would not be mali-
cious once received on the SQL server side. The ini-
tial explanation of the Random4 algorithm described
that user input would be salted before being saved in
the database. However, the authors extended to de-
scribe that the various database attributes also fall un-
der this technique rendering it harder for attackers to
crack the structure of the database.
Random4 algorithm was tested on 5 different ap-
plications proving a significant reduction in SQL in-
jection attacks when tautology is used, which is jus-
tifiable due to the nature of salting of input. Analyz-
ing the algorithm proposed by the work in [19], it is
clear that the encrypted input to SQL databases would
result in a significant performance hit due to longer
and inconsistent strings as well as the overhead pro-
duced by encrypting and decrypting all input and out-
put from and to the database.
The work presented in (Oksuz, 2022) attempted
a different approach to achieve a similar goal of
database anonymization in electronic health records
databases. The proposed model relied on blockchain
to store identifiable information. The paper explains
a novel approach to anonymize patient records in an
EHR database through the storage of medical records
by a centralized authority in an off-chain database
while referencing a hash accessible in the blockchain.
Referentiality of records is lost and all encounters
have associated blocks in the chain accessible through
a hash provided by the patient. The researchers
highlight that although records can only be identi-
fied through the presentation of a pseudo-identifier
through the patient, institutions are still capable of
utilizing anonymized data for analysis and research
purposes.
The model in (Oksuz, 2022) allows central
providers trusted on a chain to add and create new
blocks that can be associated with individual encoun-
ters in the chain. Untrusted entities requesting ac-
cess to clinical encounters would have to use prede-
termined random hashes to access various blocks as-
sociated with the same record or clinical significance
Novel Approach to De-Identify Relational Healthcare Databases at Rest: A De-Identification of Key Data Approach
211

to later consume hashes to query off-chain databases.
This requires patients to carry specific keys to all their
encounters – such as vaccination records – and would
not be able to verify the authenticity of the record un-
less provided with at least two blocks about the same
patient pseudo identifier.
This model is the most similar, in terms of func-
tionality and results, to the work proposed in this
work. Due to the fact that authors did not introduce
solutions to some quasi-identifiers such as encoun-
ters dates, it achieves a resting state of the database
that is considered a limited dataset by HIPAA defini-
tion, with pseudo identifiers available in a blockchain
only accessible through the respective block hashes.
However, in contrast to the work in this paper,
the model proposed by (Oksuz, 2022) requires a
resource-intensive setup and a complex integration of
off-chain and in-chain records, and due to the nature
of blockchain technologies, the performance impact
can be significant (Koushik et al., 2019). Further-
more, the model relies heavily on patients present-
ing access keys for all entities to access their records.
This can be counterproductive in an emergency room
setting where patients are unconscious, or hashes are
lost. The model also did not take into consideration
the necessity of case progression and timeline analy-
sis in research applications, which is very crucial in
many types of clinical and public health studies and
is pivotal to achieving continuity of care.
None of the above literature adopts an approach
that encrypts foreign keys to de-identify data in
a tiered database instance, which is the proposed
method in this research. All relevant work focused
on using anonymization when data is being extracted
for research and statistical analysis.
3 FRAMEWORK DESIGN
This section determines the proposed framework to
achieve a secure de-identified databaseenabling the
transformation of relational healthcare databases with
electronic personal health information into true de-
identified databases at rest.
3.1 Determination of Identifiers and
Quasi-Identifiers in an EHR
Database
This section suggests a categorization matrix for com-
mon electronic health records system attributes to de-
termine their protection level. The work scores the
sensitivity level of each attribute on the identifiability
continuum proposed by (El Emam, 2010) and deter-
mines the level of accessibility of each attribute. This
is achieved by ranking attributes based on the identi-
fiability continuum and by classifying the frequency
of the attribute appearance on various EHR screens.
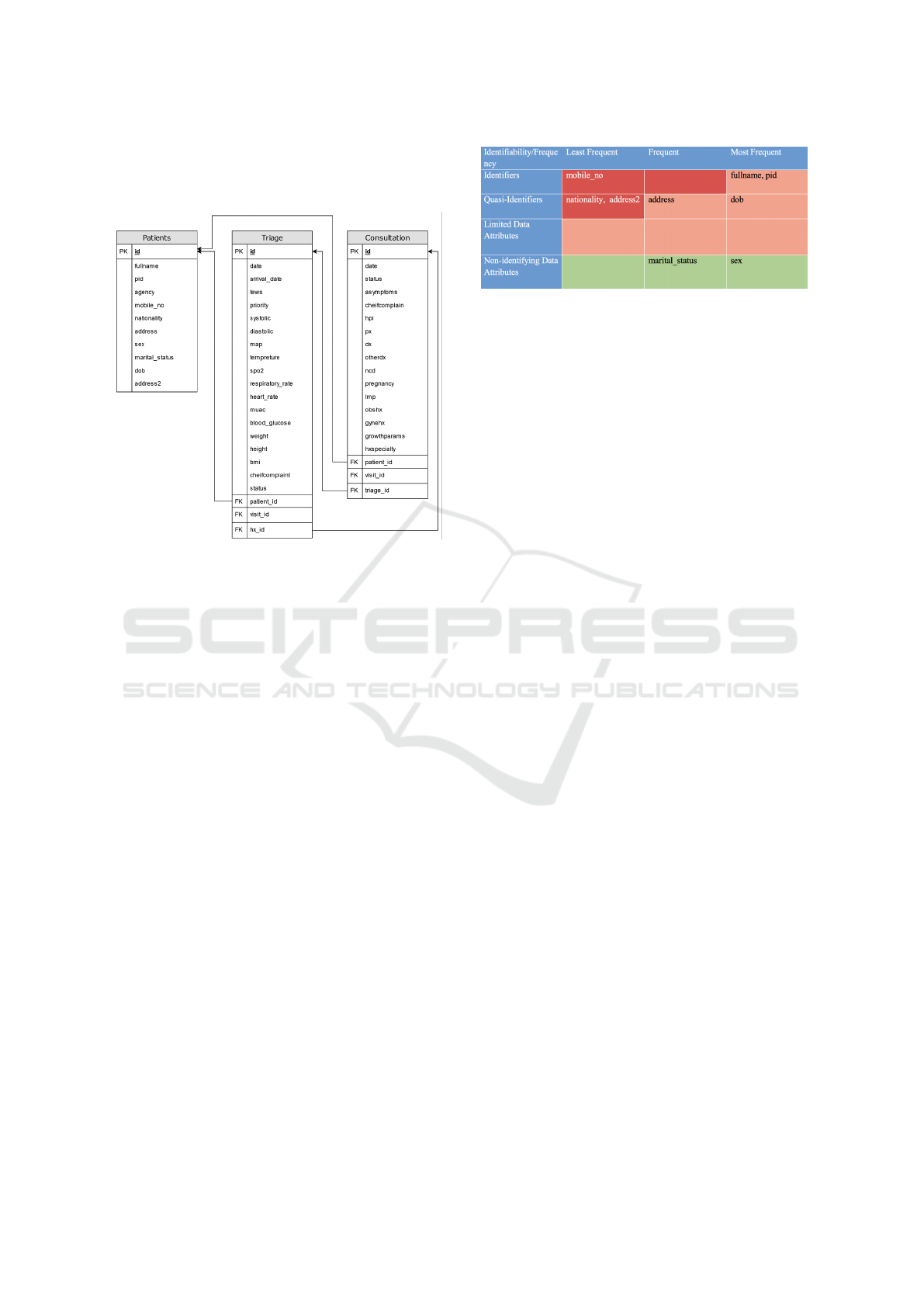
Figure 3 proposes a categorization matrix of the var-
ious data attributes based on the potential impact of
their confidentiality breach.
Figure 3: Proposed categorization of EHR attributes.
As illustrated in 3, all EHR data attributes are clas-
sified into 12 categories. Those categories offer a bal-
anced view of their sensitivity and frequency of ac-
cess. and determine their storage locations and pro-
tection policies throughout the framework. The paper
aims to classify all attributes encountered in the sam-
ple database including personal information, demo-
graphics, consultation information, and triage data.
The above categories fall in three protection levels
from highest to lowest priority as follows:
• Extremely Protected: this includes least frequent
identifiers, frequent identifiers, and least frequent
quasi-identifiers.
• Protected: this includes most frequent identifiers,
frequent quasi-identifiers, most frequent quasi-
identifiers, and all limited data attributes.
• Unprotected: all non-identifying data attributes.
The level of protection governs the level of stor-
age for the associated data attributes in the framework
database. The higher the level of storage is the more
iterations of foreign key decryptions needed to re-
trieve the data attribute as illustrated in the next chap-
ters.
3.2 Designing the Hierarchical
Structure of the Framework and
Determination of Quasi-Identifiers
Masking Methods
This section aims to outline the conceptual model of
the proposed framework. It suggests the 7 rules gov-
erning the design principles based on the protection
HEALTHINF 2025 - 18th International Conference on Health Informatics
212

level of the data attributes. Furthermore, the frame-
work considers the masking of some quasi-identifiers
such as dates while preserving the epidemiological
and clinical value.
The proposed framework assumes the preserva-
tion of the below rules:
3.2.1 Each Searchable Identifiable Attribute
Must Be Stored in a Separate Database
Table. this Eliminates the Possibility of
Cross Referencing One Identifying
Information Using Another
For searchable identifiable attributes, such as names
and unique identifiers, the proposed framework re-
quires the storage of no more than one in a single
database table. This ensures that attackers do not
get access to one identifying information because of
their previous knowledge of another (i.e., obtaining
a social security number using the patient’s name).
The highest-level table has only one encrypted for-
eign key to the next lower-level table. Lower-level
tables would have an encrypted foreign key for the
next lower-level record and another encrypted foreign
key for the previous higher-level table. The decryp-
tion of foreign keys within the software algorithm is
necessary to enable querying the next table in either
direction. Policymakers can choose whether to hier-
archically position those tables in a chained format or
to undergo grouping of attributes in various levels to
improve performance.
3.2.2 Non-Searchable Identifiable Attributes
Must Be Stored Encrypted in a Separate
Table
The proposed framework suggests that all non-
searchable identifiable attributes are stored in an
encrypted format within a separate database table.
No database table should contain columns of both,
searchable and non-searchable data attributes at any
time. Developers and security policymakers could
define whether one database table contains all non-
searchable identifiable attributes or choose to segre-
gate each attribute or group of attributes in a different
table depending on their relation to each other. Sep-
arating attributes in different tables renders the pro-
posed framework more resistant to attacks, increases
security, and further dilutes the potential harm of
database attacks. One important aspect is that the
more complex the model adopted, the less perfor-
mance is anticipated.
3.2.3 Non-Identifiable Searchable and
Non-Searchable Attributes Should Be
Stored in the Reference Database Table
This framework suggests that non-identifiable at-
tributes should be stored based on common database
structures practice. Encryption is not a necessity
for the attributes. However, the table contains the
encrypted foreign key for the lowest level identify-
ing table in the framework. This table is called the
reference table in this work. This table’s primary
key can be referenced by a foreign key in various
clinical encounters utilizing relational database fea-
tures. The proposed framework ensures that the im-
pact on performance is kept to the minimum when
using aggregate functions for the analysis purposes
of deidentified health and administrative information.
Therefore, reverse identification is possible besides
regular storage of non-encrypted attributes in the ref-
erence table. The proposed framework leaves the
structuring and normalizing of database decisions for
the system analysts and database administrators.
In case of migration to this model, analysts and
database administrators need minimal changes to
database tables below the reference table to address
quasi-identifiers such as clinical encounter dates and
caregiver information. Patient identification tables are
replaced by the reference table and database integrity
should not be affected assuming that the database has
a reasonably normalized design. This is because the
identification of patients and personnel only occurs at
one table and health information and encounters refer
to identification tables through a foreign key.
Developers and security policymakers may
choose to enhance security further and reduce the
impact of database attacks by encrypting non-
identifying attributes. However, this could have a
significant performance impact on reporting and
application data aggregation power. For this paper,
such extra encryption is out of scope and could be
revisited in future work.
3.2.4 Health Encounters and Administrative
Records Can Only Be Referenced Through
Reference Tables Producing Limited
Datasets
This approach requires that health encounters, admin-
istrative records, or any record protected under the
HIPAA privacy rule or the security policy of the or-
ganization or the region should contain a foreign key
-when needed- to the reference table containing non-
identifiable information. At this point, any identifiers
and quasi-identifiers, except encounters and admin-
istrative records dates, would be stored in a higher-
Novel Approach to De-Identify Relational Healthcare Databases at Rest: A De-Identification of Key Data Approach
213

level table above the reference table. Querying these
attributes would require decrypting one or more en-
crypted foreign keys through the application backend.
Furthermore, the resting state of the database instance
is de-identified and any direct SQL injection attack or
any other query that could be executed on the database
server is not able to retrieve any electronic personal
health information. However, it would be able to re-
trieve dated electronic health information, which is a
limited dataset. This is addressed in the next rule.
3.2.5 Dates Must Be Masked Through a
Random Increment or Decrement that Is
Different for Each Patient
To achieve a true de-identified state, dates should be
masked as they are considered quasi-identifiers and
could be utilized by attackers with previous knowl-
edge of encounter dates. The framework proposes a
random increment or decrement that is considered to
the framework as a non-identifying non-searchable at-
tribute. As discussed in design principle 3, this inter-
val is stored in a separate database table or with other
non-searchable identifiable attributes. Such a static
interval per patient would ensure visibility to proper
clinical case timeline, physicians can still observe the
progression of clinical cases on realistic timelines. A
consideration when developing the interval determi-
nation algorithm should be the fact that seasonality
and endemic information could be disrupted while
they are of high clinical significance. To overcome
this, policymakers can allow caregivers to re-produce
limited datasets or re-identify information while ad-
hering to the health information security and privacy
that apply to their organizations, for example, the
HIPAA privacy rule requires logging any access to
limited or identified datasets.
3.2.6 Incremental Primary Keys Are not
Allowed for any Table Above the
Reference Table
With incremental primary keys, attackers can infer the
relation between two tables through a sequence, an
attacker might relate that entry number 1 in the pa-
tient names table refers to entry number 1 of the social
security numbers table. To ensure maximal security,
and to inhibit the ability of attackers to infer personal
health information based on the sequence of insert
and creation or timestamps. The proposed framework
requires that incremental primary keys be not allowed
for any of the database tables above the reference ta-
ble. As an example, developers can use universally
unique identifiers (UUID) for tables without a unique
attribute.
3.2.7 Encryption Keys Should Be Different for
Every Encrypted Foreign Key
Developers and policymakers achieve a higher level
of protection when encryption keys are different for
each foreign key, this ensures that if any foreign key
is jeopardized through the software interfaces, the ap-
plication tier, or improper key management, other for-
eign keys would still be protected and immune to the
attack and the data leak would be to a minimum. Key
management methods and procedures are out of scope
for this work but could be examined in future work.
Figure 4: Proposed schema of EHR data attributes.
Adopting the schema proposed in Fig. 4 through
implementing the 7 rules proposed in this section
would result in loss of referential integrity between
the various tables above the reference table. This is
caused the encrypted foreign keys that are handled the
application layer. This referential integrity loss was
observed in all methodologies segregating identifiers
and quasi-identifiers from health encounters. How-
ever, unlike other proposed work examined in this
work, this framework sustains a one-to-one relation-
ship between all tables above the reference table with
no change to referential integrity with the clinical en-
counters. This simplifies implementing integrity con-
straints at the application layer to compensate for the
referential integrity loss anticipated by adopting this
framework.
3.3 Applying the Framework to an
EHR Database
This section aims to apply this framework to a con-
ceptual EHR database, during this phase a typical
SQL database schema for an electronic health records
system is used to adapt the framework, the software
database is structured to have 70 different tables that
cover primary, secondary, and tertiary health care ser-
vices. For this study, we de-identify the database ta-
HEALTHINF 2025 - 18th International Conference on Health Informatics
214
bles that store only primary health records, admin-
istration records, and patient identification tables to
provide an example for a wider use.
Figure 5: Original database structure.
Figure 5 shows the original structure of the
database. Three database tables are de-identified us-
ing the framework proposed structure for this work.
• Patients Information Table: This table contains
identifiers and quasi-identifiers of all patients pro-
cessed through the software.
• Triage Information Table: This table contains in-
formation from various nursing stations in pri-
mary healthcare settings, the information stored in
this table are the patient’s vital signs, chief com-
plaints, and a few administrative attributes along
with service received dates.
• Clinical Consultations Table: This table contains
the consultation information of all cases. It stores
all consultation details as well as the diagnosis
and the treatment plan of the patient, which are
the most sensitive information to be protected.
To restructure the schema presented in Figure 5
abiding to the seven design principles outlined previ-
ously in this work, a categorization of data attributes
need to take place based on the frequency and iden-
tifiably of the attributes. This categorization helps
in identifying the correct level to store each attribute
in the new schema. We concluded the categoriza-
tion highlighted in Figure 6 of patient personal in-
formation attributes based on typical electronic health
records systems functionality.
Given the encryption of foreign keys in the ap-
plication database, it is anticipated that the database
Figure 6: Attributes categorization using the identifiability
continuum.
loses referential integrity between the higher-level ta-
bles. Software developers can implement various
constraints at the application layer to ensure referen-
tial integrity between those tables.
3.3.1 Step 1: Searchable Identifiable Attributes
Must be Stored in a Separate Database
Table
To adapt to the first rule, identifiable information
that is searchable “Non-Encrypted” would have to be
stored in separate tables to preserve confidentiality.
This ensures that an attacker gaining access to the
database would not be able to infer, or correlate iden-
tifiers based on previous knowledge of one or more
of those identifiers. An example would be an attacker
aware of a specific patient’s name being able to re-
trieve their social security number due to their storage
in one row. Both the patient’s full name and patient
unique identifier attributes highlighted in Figure 6 are
considered searchable (frequent) identifiers. To abide
by the framework rules, they are stored in separate
database tables both with encrypted foreign keys re-
lating to the next level table. Developers can choose
to hierarchically structure those tables as separate lev-
els of this framework or store the various attributes on
the same level. If a similar level is chosen develop-
ers must use different encryption keys or vectors for
both tables. This ensures that the encrypted foreign
key value is not similar in both tables and cannot be
used for reference.
Figure 7 illustrates the implementation of the
framework for searchable identifiable attributes while
storing each attribute in a separate level. This repre-
sents a more secure design in comparison to Figure 8.
However, this design requires back-end functions to
iterate through subsequent levels to arrive at the ref-
erence table used to relationally reference the clini-
cal encounters resulting in performance degradation.
This work uses this design in the database transforma-
tion and all results are shared to the most secure im-
plementation of this framework. Developers and or-
ganizations can weigh the security benefits in compar-
ison to the performance impact to determine whether
Novel Approach to De-Identify Relational Healthcare Databases at Rest: A De-Identification of Key Data Approach
215
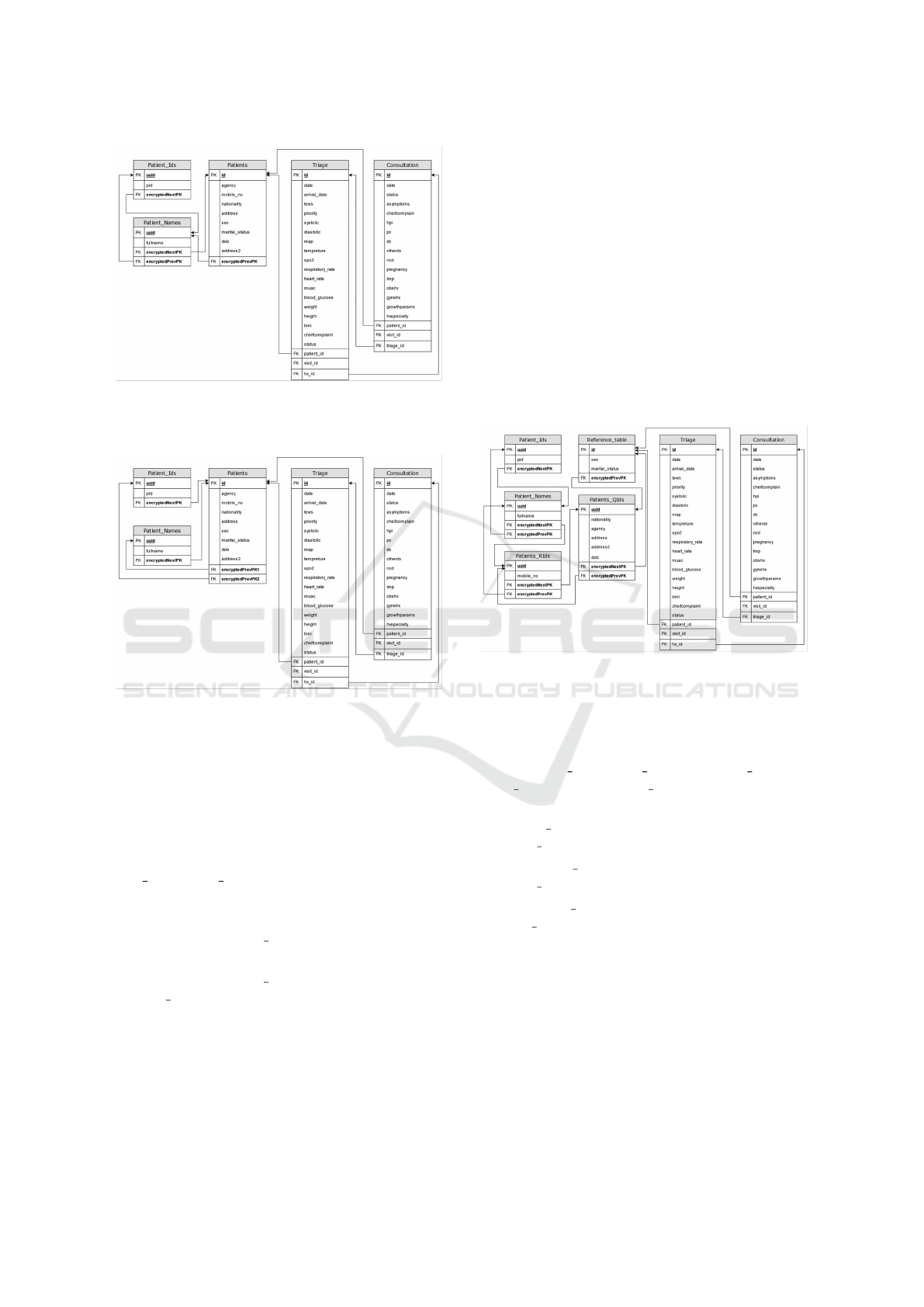
Figure 7: Searchable identifiable attributes must be stored in
a separate database table for each attribute, separate levels
design (more secure).
Figure 8: Searchable identifiable attributes must be stored
in a separate database table for each attribute, same level
design (less secure).
to add their searchable identifiable attributes in one or
multiple levels.
To ensure that users with access to the database
are unable to utilize relational features to infer ePHI
or any other identifiable information. The encryption
key or encryption vector should be different for Pa-
tient Ids, Patient Names, and Patients tables. To en-
sure proper adoption of the seventh rule of the frame-
work, the following rules must be true:
• In Figure 7: Patient Ids.encryptedNextPK ̸= Pa-
tients.encryptedPrevPK.
• In Figure 8: Patient Ids.encryptedNextPK ̸= Pa-
tient Names.encryptedNextPK.
3.3.2 Step 2: Non-Searchable Identifiable
Attributes Must be Stored Encrypted in a
Separate Table
To abide by this rule, all identifiers and quasi-
identifiers that are non-searchable must be removed
from the reference table and preferably stored in an
encrypted format. As discussed previously, policy-
makers can choose whether to store both identifiers
and quasi-identifiers at the same level or not as well
as the possibility of grouping those attributes accord-
ing to their relevance and the cross-matching possibil-
ity of re-identification. For example, if the insurance
identification number is stored in the same table with
the name or id of the insurance provider, the possibil-
ity of inference increases if attackers gain access to
information in that specific table. Policymakers can
choose to store those two attributes in separate levels
of the framework to increase security keeping in mind
the potential impact on performance. Both implemen-
tations are illustrated in Figures 9 and 10.
Figure 9: Non-searchable identifiable attributes must be
stored encrypted in a separate table. separate levels design
(more secure).
To prevent violation of the framework rules, the
encryption key or encryption vector should be differ-
ent for Patient Ids, Patient Names, Patients RIds, Pa-
tients QIds, and Reference table tables. This results
in the following rules to be true:
• Patient Ids.encryptedNextPK ̸= Pa-
tients RIds.encryptedPrevPK.
• • Patient Names.encryptedNextPK ̸= Pa-
tients QIds.encryptedPrevPK.
• • Patient RIds.encryptedNextPK ̸= Refer-
ence table.encryptedPrevPK.
Although Figure 10 illustrates a higher-
performing design, it is less secure than the model
illustrated in Figure 9.
3.3.3 Step 3: Non-Identifiable Searchable and
Non-Searchable Attributes Should be
Stored in the Reference Database Table
As discussed in the previous section, successful im-
plementation of rules 1, 2, and 7 from the frame-
work results in only non-identifiable information to
HEALTHINF 2025 - 18th International Conference on Health Informatics
216
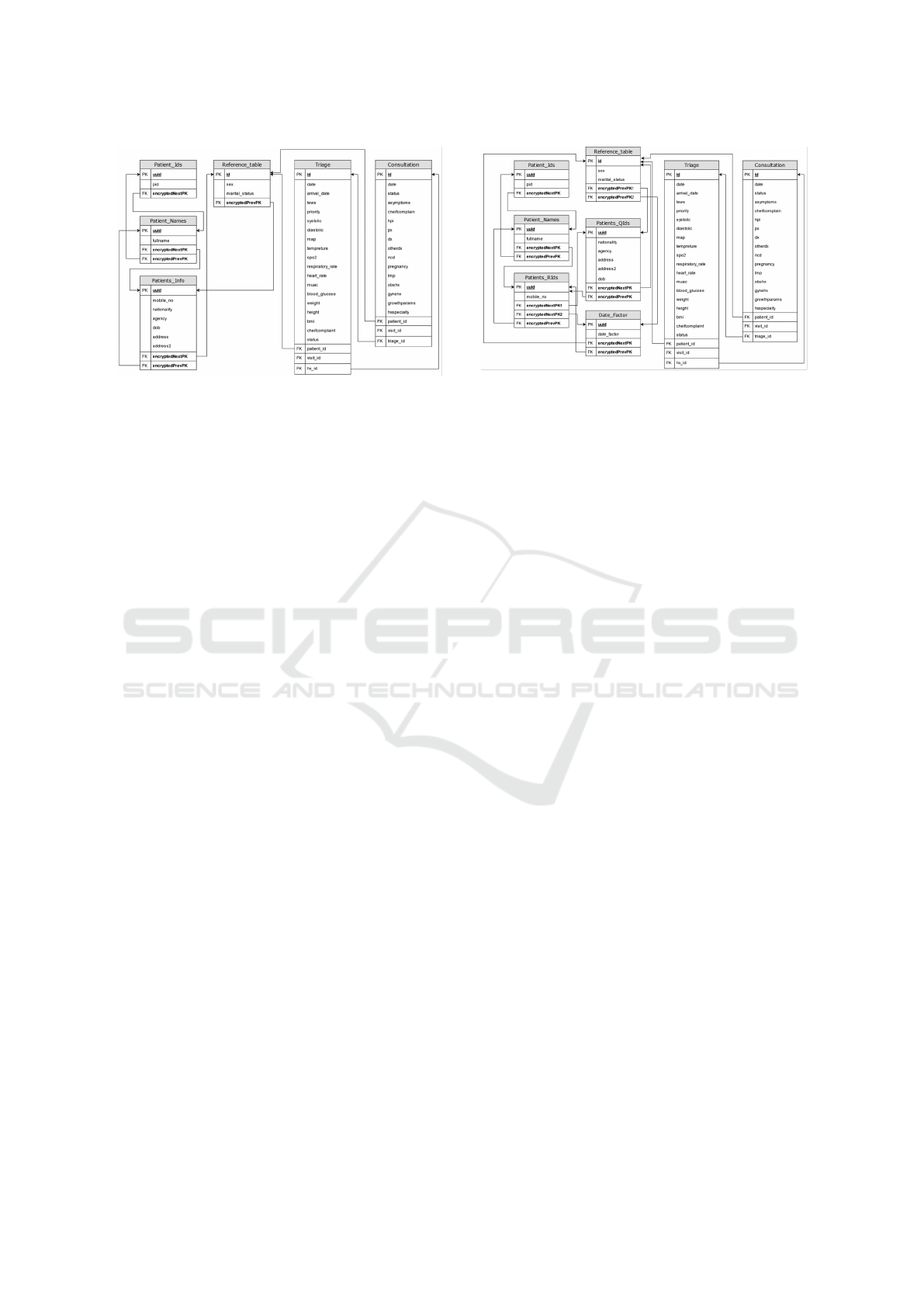
Figure 10: Non-searchable identifiable attributes must be
stored encrypted in a separate table. Single table design
(less secure).
be stored in the reference table. This allows EHR
solutions to leverage relational database features to
process and manipulate data stored within the ref-
erence table without any decryption required by the
backend application or any change in the application’s
logic. Researchers continue to have the ability to
leverage EHR data for public health, disease surveil-
lance, and other research purposes. This is because
the framework preserves non-identifiable information
in the reference table. More complex research stud-
ies such as time series and case studies would require
specific algorithms to be implemented in the back-end
application.
3.3.4 Step 4: Dates Must be Masked Through a
Random Increment or Decrement that is
Different for Each Patient
To achieve a higher level of security for databases in
their resting state, dates and timestamps are masked
as they are considered quasi-identifiers. Attackers can
infer patients’ identifiable information if they have
pre-existing knowledge of clinical encounters’ time
stamps. To achieve this, this model proposes a ran-
dom increment or decrement automatically assigned
to each patient at the registration stage. This factor
is used to mask all records associated with the pa-
tients in all tables regardless of their location within
the framework. The random static factor ensures that
data integrity and quality for researchers remain intact
for case progression and time series analysis. How-
ever, seasonality might be lost which could be tackled
by using a less secure year increment/decrement fac-
tor. Given that dates are considered quasi-identifiers
in health applications, the factor is stored at a similar
level with other quasi-identifiers.
Figure 11 illustrates the final proposed database
structure implementing all rules of the proposed
Figure 11: Final Proposed database structure, abiding to all
framework rules.
framework. The main patient’s table illustrated in
Figure 3.2 was split into 5 smaller tables with en-
crypted foreign keys referring to the next and previ-
ous level tables. The framework preserved the origi-
nal patients’ identifiers in all encounters tables and re-
sulted in no change in their structure enabling a sim-
pler adoption of the framework due to the minimal
changes needed in software logic to reflect change.
The final proposed database structure stores
searchable identifiable attributes in separate tables
structured in a hierarchical manner (Rule 1). It
also stores less frequently used identifiers at a dif-
ferent level from quasi-identifiers (Rule 2). All non-
identifying attributes are stored in the reference table
for easy retrieval (Rule 3). All encounters’ tables refer
to the reference table (Rule 4) preserving all the refer-
ential features of relational databases with no change
reducing the impact on database utilization of data
aggregation and research. The structure extends to
store the date increment/decrement factor separately
for each patient within the database (Rule 5) and abid-
ing by (Rule 6) at all levels above the reference table.
The framework utilizes universally unique identifiers
(UUIDs) limiting the possibility of inference due to
relevant records ordering in all tables. When imple-
menting this structure, encryption keys or vectors are
different for different levels ensuring that relational
features of the database are not used to re-identify
data (Rule 7).
The implementation of this approach in pre-
existing EHR systems would require those changes
to be executed at tables containing patient identifiers,
with minimal to no changes at the clinical encounters
tables. For applications utilizing best practices such
as database normalization and modular application
design, the implementation of this approach would
be relatively simple and very similar to the approach
presented in this paper. Efficient utilization of this ap-
proach would highly rely on adopting an application
Novel Approach to De-Identify Relational Healthcare Databases at Rest: A De-Identification of Key Data Approach
217

design that minimizes re-identification. Example ap-
proaches would be to temporarily store de-identified
patient information in the session as providers are
navigating the relevant patient history or utilizing
object oriented architectures where re-identification
would happen once for active patient’s object.
4 CONCLUSION AND FUTURE
WORK
De-identification of medical data has been a widely
used solution to produce data extracts for research
and analysis. The work was able to identify 7 frame-
work principles that could de-identify any health in-
formation system database through Segregation of
identifiable information in separate database tables
based on their importance and frequency, omitting the
use of relational database features between those ta-
bles through encryption of foreign keys, and address-
ing quasi-identifiers such as encounters dates through
masking with a random increment/decrement that is
stored in the same manner. The de-identification of
a sample EHR schema database was successful mi-
grating the original database structure to a structure
conforming to the 7 principles of the framework.
In future work, our aim is to test the framework
on a real-life EHR database and compare the perfor-
mance against the original to determine the suggested
framework efficiency.
REFERENCES
Avireddy, S., Perumal, V., Gowraj, N., Kannan, R. S., Thi-
nakaran, P., Ganapthi, S., Gunasekaran, J. R., and
Prabhu, S. (2012). Random4: An application specific
randomized encryption algorithm to prevent sql injec-
tion. In 2012 IEEE 11th International Conference on
Trust, Security and Privacy in Computing and Com-
munications, pages 1327–1333.
Caplan, R. (2003). Hipaa. health insurance portability and
accountability act of 1996. Dental assistant (Chicago,
Ill. : 1994), 72:6–8.
Capris, T., Melo, P., Garcia, N. M., Pires, I. M., and
Zdravevski, E. (2022). Comparison of sql and nosql
databases with different workloads: Mongodb vs
mysql evaluation. In 2022 International Conference
on Data Analytics for Business and Industry (ICD-
ABI), pages 214–218.
Chhabra, S., Amiri, H., Rastegar, M., and Dashti, A. (2022).
Cloud computing for healthcare systems in covid19
era. Open Access Research Journal of Biology and
Pharmacy, 06.
El Emam, K. (2010). Risk-based de-identification of health
data. IEEE Security & Privacy, 8(3):64–67.
Enterprise, V. (2018). 2018 data breach digest report.
Erdal, B., Liu, J., Ding, J., Chen, J., Marsh, C., Kamal, J.,
and Clymer, B. (2012). A database de-identification
framework to enable direct queries on medical data for
secondary use. Methods of information in medicine,
51:229–41.
Koushik, A. S., Jain, B., Menon, N., Lohia, D., Chaudhari,
S., and B.P, V. K. (2019). Performance analysis of
blockchain-based medical records management sys-
tem. In 2019 4th International Conference on Recent
Trends on Electronics, Information, Communication
& Technology (RTEICT), pages 985–989.
Lin, J. C.-W., Liu, Q., Fournier-Viger, P., and Hong, T.-
P. (2016). Pta: An efficient system for transaction
database anonymization. IEEE Access, 4:6467–6479.
Miller, A. and Payne, B. (2016). Health it security: An ex-
amination of modern challenges in maintaining hipaa
and hitech compliance. 2016 KSU Conference on Cy-
bersecurity Education, Research and Practice.
Oksuz, O. (2022). A System For Storing Anonymous Pa-
tient Healthcare Data Using Blockchain And Its Ap-
plications. The Computer Journal, 67(1):18–30.
Omran, E., Bokma, A., and Abu-Almaati, S. (2009). A k-
anonymity based semantic model for protecting per-
sonal information and privacy. In 2009 IEEE Interna-
tional Advance Computing Conference, pages 1443–
1447.
OWASP (2024). Owasp top ten.
Patel, D., Dhamdhere, N., Choudhary, P., and Pawar, M.
(2020). A system for prevention of sqli attacks. In
2020 International Conference on Smart Electronics
and Communication (ICOSEC), pages 750–753.
Rights, P. (2023). Privacy breaches.
Sai Lekshmi, A. S. and Devipriya, V. S. (2017). An emula-
tion of sql injection disclosure and deterrence. In 2017
International Conference on Networks & Advances
in Computational Technologies (NetACT), pages 314–
316.
Singh, A. (2024). Evolutionary architectures in web ap-
plications: A comprehensive study of client-server,
multi- tier, and service-oriented approaches. IJFMR.
Unlu, S. A. and Bicakci, K. (2010). Notabnab: Protection
against the “tabnabbing attack”. In 2010 eCrime Re-
searchers Summit, pages 1–5.
HEALTHINF 2025 - 18th International Conference on Health Informatics
218
