
Deep Learning for Image Analysis and Diagnosis Aid of Prostate Cancer
Maxwell Gomes da Silva
1
, Bruno Augusto Nassif Travenc¸olo
1
and Andr
´
e R. Backes
2
1
School of Computer Science, Federal University of Uberl
ˆ
andia, Uberl
ˆ
andia, Brazil
2
Department of Computing, Federal University of S
˜
ao Carlos, S
˜
ao Carlos-SP, Brazil
Keywords:
Deep Learning, Prostate Cancer, Image Segmentation.
Abstract:
Prostate cancer remains one of the most critical health challenges, ranking among the leading causes of cancer-
related deaths in men worldwide. This study seeks to automate the identification and classification of cancer-
ous regions in histological images using deep learning, specifically convolutional neural networks (CNNs).
Using PANDA dataset and Mask R-CNN, our approach achieved an accuracy of 91.3%. This result highlights
the potential of our methodology to enhance early detection, improve patient outcomes, and provide valuable
support to pathologists in their diagnostic processes.
1 INTRODUCTION
Prostate cancer represents a major global health chal-
lenge, ranking among the leading causes of cancer-
related mortality in men, with millions of new cases
diagnosed annually. In Brazil, it is the fourth lead-
ing cause of cancer deaths, accounting for 6% of all
cancer-related fatalities. Reports from 2022 indicate a
concerning increase, with approximately 71,730 new
cases and 16,301 deaths, representing 29.2% of male
cancer cases Humphrey (2004). Similarly, in the
United States, 2022 saw around 288,300 new cases
and 34,700 deaths, with projections indicating that 1
in 8 men will face a prostate cancer diagnosis during
their lifetime Society (2023).
In response to the rising incidence and mortal-
ity rates, Brazil and the United States have imple-
mented comprehensive cancer prevention and con-
trol policies. These initiatives focus on raising pub-
lic awareness of risk factors, promoting early detec-
tion, and ensuring equitable access to quality treat-
ment—critical steps in reducing the disease’s burden
Humphrey (2004); Society (2023).
Early detection of prostate cancer, typically
achieved through histopathological analysis of biopsy
tissue, is pivotal for effective treatment. However, this
process is inherently subjective, depending on pathol-
ogists’ expertise, and prone to variability. These lim-
itations underscore the need for computational tools
to enhance diagnostic precision and efficiency. Ad-
vances in artificial intelligence, particularly in con-
volutional neural networks (CNNs) and instance seg-
mentation models like Mask R-CNN, offer promising
avenues for automating the identification and classifi-
cation of histological patterns.
This study aims to develop a comprehensive
methodology for diagnosing prostate cancer through
image processing and analysis techniques. Our ap-
proach involves curating a dataset of whole slide his-
tological images and segmenting and classifying each
image based on Gleason Scores. We trained a CNN
and evaluated its performance in detecting and classi-
fying prostate cancer.
The remainder of this paper is organized as fol-
lows: Section 2 outlines the materials and methods
used in this study. Section 3 reviews related work.
Section 4 details our prostate cancer classification ap-
proach based on Gleason Scores. Section 5 presents
and discusses the results. Finally, Section 6 concludes
the paper.
2 THEORETICAL BACKGROUND
2.1 Prostate Cancer and Gleason Score
Prostate cancer is a prevalent malignancy that affects
the prostate gland, a small organ situated below the
bladder and in front of the rectum in men. The dis-
ease occurs when cells in the prostate grow uncon-
trollably, forming tumors that may eventually spread
to other parts of the body. Risk factors include age,
family history, and certain genetic mutations. While
prostate cancer often progresses slowly, some forms
Gomes da Silva, M., Travençolo, B. A. N. and Backes, A. R.
Deep Learning for Image Analysis and Diagnosis Aid of Prostate Cancer.
DOI: 10.5220/0013302400003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 3: VISAPP, pages
699-706
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
699

can be aggressive and advance rapidly, making early
detection critical for effective management and treat-
ment.
Diagnosis typically involves a histopathological
examination of prostate biopsy tissue, recommended
when abnormalities are identified through digital rec-
tal examinations or elevated Prostate-Specific Anti-
gen (PSA) levels Loeb et al. (2014). The Gleason
score, derived from histopathological assessments,
evaluates the cancer’s histological grade, aiding in
predicting tumor growth rate, and metastatic poten-
tial, and guiding patient treatment plans Brazil (2002).
The National Cancer Institute classifies prostate
cancer on a Gleason grading scale ranging from 1 to
5 Humphrey (2004):
• Grade 1 - Cells are uniform and small, forming
regular glands with minimal variation in size and
shape. The margins are well-defined, and cells are
densely clustered with minimal stroma between
them.
• Grade 2 - Cells exhibit more variation in size and
shape, though the glands remain relatively uni-
form. Nodules are loosely arranged with irregular
borders.
• Grade 3 - Cells display greater variability in size
and shape, forming small, irregularly distributed
glands that may be angled or elongated. Spindle-
shaped or papillary nodules with smooth borders
may also be present.
• Grade 4 - Many cells merge into large, amorphous
masses or irregular glands unevenly distributed.
Signs of infiltration and invasion into adjacent tis-
sues are apparent.
• Grade 5 - Tumor cells are anaplastic, aggregating
into large clumps that invade nearby organs and
tissues. Central necrosis may be observed, often
with a comedocarcinoma pattern. Glandular dif-
ferentiation is frequently absent, and growth may
appear cord-like or loosely arranged, indicating
infiltration.
The diagnostic process relies on analyzing biopsy
tissue images and assigning a Gleason score to clas-
sify the histological grade of the tumor. Currently,
this analysis is performed manually by specialists—a
time-intensive process prone to human error. How-
ever, with advancements in cancer diagnostic tech-
nologies, there is significant potential to develop al-
gorithms capable of identifying cancer-prone regions
and classifying them based on the Gleason score. Ex-
isting algorithms have shown promise, achieving ac-
curacy rates exceeding 77% in cancer classification
based on known disease characteristics from medical
reports Bulten et al. (2022).
2.2 Mask R-CNN for Instance
Segmentation
An Artificial Neural Network (ANN) is a highly par-
allel, distributed processor comprising simple pro-
cessing units that store experimental knowledge and
make it accessible for practical applications Ro-
drigues et al. (2022). Similar to the human brain,
an ANN acquires knowledge from its environment
through a learning process and stores this knowledge
in the connections between neurons Haykin (1998).
Extending this concept, the Convolutional Neural
Network (CNN) draws inspiration from the brain’s hi-
erarchical feature-learning mechanisms Ghose et al.
(2012).
A CNN is structured around three primary types
of layers:
• Convolutional Layer: Performs convolution oper-
ations to extract feature maps.
• Pooling Layer: Reduces the spatial dimensions of
feature maps while retaining critical information,
improving computational efficiency, and reducing
overfitting.
• Fully Connected Layers: Transforms feature
maps into a format suitable for classification, al-
lowing the network to categorize input data into
different classes Kang and Wang (2014).
Instance segmentation, a crucial challenge in com-
puter vision, involves accurately identifying and lo-
calizing objects within an image at the pixel level. A
leading approach for this task is the Mask R-CNN
algorithm, a state-of-the-art neural network specifi-
cally designed for object detection and segmentation.
Mask R-CNN excels in distinguishing multiple ob-
jects within an image and generating bounding boxes
and pixel-level masks for each Chiao et al. (2019).
Mask R-CNN operates using a two-stage frame-
work:
• Proposal Generation: Identifies Regions of Inter-
est (ROIs) within the input image where objects
of interest may exist.
• Detailed Refinement: Refines these proposals
by predicting object classes, adjusting bounding
boxes, and generating precise pixel-level masks
for each ROI Gonzalez and Woods (2008).
In this study, we used the Mask R-CNN structure
implemented within the Detectron2 framework Wu
et al. (2021), comprising the following components:
1. Backbone: A Convolutional Neural Network
(ResNet) is employed to extract features from in-
put images. ResNet’s architecture incorporates
residual blocks, enabling the training of deeper
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
700

networks with improved accuracy and reduced
vanishing gradients.
2. Bottleneck Blocks: These building units of
ResNet contain multiple convolutional layers, of-
ten incorporating shortcuts to streamline informa-
tion flow. The bottleneck blocks enhance feature
extraction across various abstraction levels.
3. Regions of Interest (ROI): After feature extraction
by the backbone, the model identifies specific re-
gions within the image that may contain objects
of interest. These regions are marked for further
analysis.
4. Prediction Heads: Dedicated prediction heads are
used to perform distinct tasks within the ROIs:
• Class Prediction: Identifies the class of objects.
• Bounding Box Prediction: Determines the co-
ordinates of bounding boxes surrounding de-
tected objects.
• Mask Prediction: Generates pixel-level masks
that delineate the precise boundaries of each
object.
These components synergistically enable Mask R-
CNN to perform object detection and segmentation,
from initial feature extraction to precise object iden-
tification and localization within images. This capa-
bility makes it a powerful tool for complex computer
vision tasks.
2.3 Dataset
In our work, we used the PANDA dataset, a collec-
tion of prostate cancer biopsy images. The images
in this dataset vary in size, ranging between 21 and
50 megabytes on average, with typical dimensions of
8,192 pixels in width by 22,528 pixels in height. They
are 24-bit color images in TIFF format, requiring a to-
tal storage space of 411.9 gigabytes Kaggle (2023).
This dataset is divided into two subsets. The first
subset, named Radboud, contains histological images
of prostate glands with detailed annotations for indi-
vidual tissue types, categorized as follows: stroma
(connective tissue or non-epithelial tissue); healthy
epithelium (benign epithelial tissue); cancerous ep-
ithelium (Gleason 3); cancerous epithelium (Gleason
4); and cancerous epithelium (Gleason 5). The second
subset, Karolinska, provides broader region-level la-
bels, defined as background, non-tissue, or unknown
regions; benign tissue, a combination of stroma and
epithelial tissue; and cancerous tissue, a combination
of stroma and epithelial tissue exhibiting malignancy
Kaggle (2023).
For this study, we focused on the Radboud sub-
set due to its inclusion of additional metadata, such
as segmentation masks for each image. These masks
provide Gleason Score classifications and highlight
specific regions indicating the presence of prostate
cancer, offering finer granularity essential for our
analysis and model training.
3 RELATED WORK
In recent years, numerous automatic segmentation
methods for prostate imaging in magnetic resonance
imaging (MRI) have been proposed, playing a crit-
ical role in prostate cancer management, including
detection, biopsy, staging, monitoring, and treatment
Brazil (2002); Toth et al. (2014). One such approach,
presented in LeCun et al. (2010), relies on atlas-based
region matching using contours, achieving a mean
Dice Similarity Coefficient (DSC) of 84.4% on the
PROMISE12 MRI dataset. Similarly, the study in
Tian et al. (2015) utilized multiple atlases, incorpo-
rating prostate volume and contour, to refine initial
segmentations. By selecting the most similar atlas
to the segmented image, the method reached a mean
DSC of 84.0% on the same dataset. Another strat-
egy employed superpixels combined with a Random
Forest classifier for prostate segmentation, achiev-
ing a DSC accuracy of 88.0% on the PROMISE12
dataset Humphrey (2004). A hierarchical grouping
approach with statistical analysis, described in Yan
et al. (2016), obtained an impressive DSC rate of
92.05% on a private database.
These methods are generally categorized into four
types: contour-based, region-based, classification-
based (either supervised or unsupervised), and hy-
brid methods Ghose et al. (2012). Each has dis-
tinct advantages and challenges. For instance, region-
based methods are intuitive but require manual pa-
rameter adjustments, while contour-based methods
adapt quickly yet are sensitive to variations in prostate
shape. Classification-based methods are accurate and
fast but demand a robust training dataset and care-
ful feature selection Ghose et al. (2012); Tian et al.
(2015); Aldoj et al. (2020).
Deep learning, particularly Convolutional Neural
Networks (CNNs), has emerged as a transformative
approach, surpassing traditional methods by directly
learning features and patterns from images. In Chiao
et al. (2019), a deep learning-based system for Glea-
son grading in prostate cancer biopsy images achieved
77% accuracy on the SICAPv2 dataset. Similar deep
learning advancements have been observed in other
medical imaging domains, including breast cancer
detection through ultrasound imaging Chiao et al.
(2019) and oral cancer diagnosis from histological
Deep Learning for Image Analysis and Diagnosis Aid of Prostate Cancer
701

Figure 1: Example of a histological section of tissue where
morphological operations were applied. The areas outlined
in pink (A) represent the regions where these operations
were performed, resulting in the removal of certain struc-
tures. The areas outlined in black (B) represent the regions
that were preserved and considered for calculating the Glea-
son Score.
images dos Santos et al. (2023, 2021). These develop-
ments underscore the growing efficacy of automated
systems in enhancing diagnostic precision and effi-
ciency across medical imaging tasks.
4 EXPERIMENTAL SETUP
To prepare the images for analysis, we began by per-
forming morphological operations aimed at noise re-
duction, eliminating objects smaller than 100 pix-
els, and filling in gaps within objects up to 25 pix-
els in size Lu et al. (2019). Subsequently, we gen-
erated masks by segmenting image annotations and
assigning distinct colors to represent various cate-
gories: background (black), stroma (gray), healthy
epithelium (blue), cancerous epithelium with Gleason
Grade 3 (yellow), cancerous epithelium with Gleason
Grade 4 (red), and cancerous epithelium with Glea-
son Grade 5 (green). These masks were instrumental
in delineating object boundaries and excluding empty
regions from the dataset. Figure 1 illustrates the out-
comes of these morphological operations and the re-
finement of segmentation.
The morphological operations were pivotal in im-
proving the quality of image masks by enhancing ob-
ject contours and removing irrelevant components.
We applied a small-object removal process to filter
out regions with areas below a predetermined thresh-
old, effectively eliminating noise and artifacts that
could hinder the analysis. Following this, the remain-
ing connected regions were reprocessed and labeled
for further analysis. To visually emphasize signifi-
cant regions, we extracted and overlaid contours onto
the original images, providing a clear representation
of segmented areas and their adjustments. This step,
as demonstrated in Figure 1, was essential for refin-
ing the segmentation and ensuring the accuracy of the
dataset.
The processed images and masks were then di-
vided into patches measuring 512 × 512 pixels. We
retained only patches containing objects defined by
the masks. To ensure a balanced dataset, we identified
the category with the fewest items and randomly se-
lected additional items from other categories to match
this number. This resulted in a total of 28,309 sam-
ples, with each category comprising 5,661 items. Fig-
ure 2 depicts the final output of the preprocessing
stage, illustrating the uniform distribution of samples
across categories.
We structured the resulting database in the COCO
(Common Objects in Context) format, which is
widely used in machine learning for object classifi-
cation and segmentation tasks Tsung-Yi Lin (2015).
This format facilitated the creation of classification
categories corresponding to Gleason Scores and re-
gions, alongside masks for training convolutional
neural network (CNN) models.
For segmentation, we employed the Mask R-
CNN implementation from Detectron2, using lizing
a ResNet-50 backbone pre-trained on ImageNet Kyle
and Hricak (2000). The training process involved a
batch size of 12, AdamW optimizer and a learning
rate of 0.0001. We allocated 80% of the images for
training and 20% for validation. We conducted train-
ing over 40,000 iterations (approximately 22 epochs),
with learning rate reductions applied at 70% and 90%
of the training duration, halving the learning rate at
these points to optimize convergence.
Hyperparameter choices were based on both ex-
perimental results and established literature, aiming
to tailor the model’s performance to our specific
dataset. Initially, we employed Detectron2’s default
settings and subsequently loaded a pre-configured file
containing parameters optimized for our task Wu et al.
(2019); He et al. (2016); Szegedy et al. (2016).
To maintain a balanced dataset, we randomly se-
lected images and their corresponding masks across
all Gleason Score categories, ensuring the number of
samples in each category matched the one with the
fewest samples. This approach allowed us to create
a homogeneously distributed dataset, essential for un-
biased model training.
Quantitative metrics are essential to assess model
performance and guide subsequent adjustments.
These metrics measure, compare, and track model
performance, and allow for the evaluation and con-
tinuous improvement of algorithms. In our study, we
used four main metrics to assess model behavior and
generalization ability. They are:
Accuracy: measures the proportion of correct predic-
tions relative to the total number of predictions:
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
702

Figure 2: Example of preprocessing: A) Original image; B) Mask detected through annotations available in the dataset; C)
Mask segmented into regions with objects; D) Original image segmented by the same regions as its mask.
Accuracy =
Number of correct predictions
Total number of predictions
(1)
False Negative Rate: is the proportion of actual in-
stances of a class that were incorrectly classified as
not belonging to the class:
FNR =
False Negative
True positives + False Negative
(2)
Classification Loss: measures the difference be-
tween the model’s class predictions and the actual
classes. It is usually calculated using cross-entropy:
Classification Loss = −
N
∑
i=1
y
i
log( ˆy
i
) (3)
where y
i
is the actual class and ˆy
i
is the predicted
probability for the class i.
Mask Loss: measures the difference between pre-
dicted and actual binary masks, and is used to adjust
the accuracy of object segmentations in images:
L
mask
= BCE(
ˆ
M, M) (4)
where L
mask
represents the “Mask Loss”, BCE is the
abbreviation for “Binary Cross-Entropy Loss”,
ˆ
M is
the predicted mask and M is the ground truth mask.
5 RESULTS AND DISCUSSION
Figure 3 illustrates the model’s accuracy throughout
its training process, which is a key metric reflect-
ing the model’s ability to correctly classify images.
This ability is essential for ensuring the reliability of
the system in practical clinical applications. In our
study, we achieved a significant accuracy of 91.34%
in identifying prostate cancer within the dataset. Fur-
thermore, the features learned by the network demon-
strated strong generalization capabilities, with only
a 0.68% difference in accuracy between the training
and validation datasets.
Figure 4 shows the percentage of false negatives
identified by the model. The false negative rate is
Deep Learning for Image Analysis and Diagnosis Aid of Prostate Cancer
703
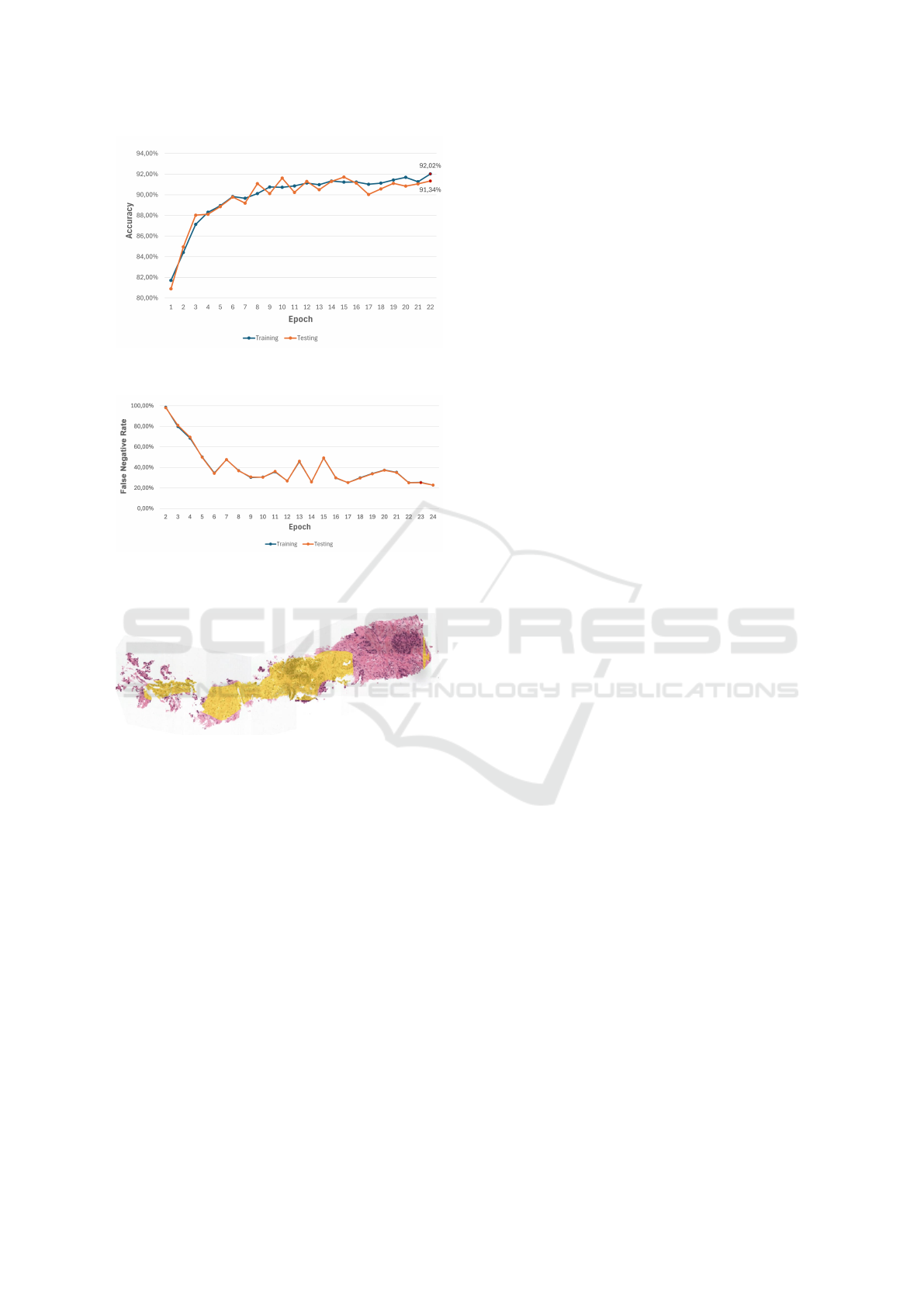
Figure 3: Accuracy of the model throughout its training pro-
cess.
Figure 4: False Negative Rate of the model throughout its
training process.
Figure 5: Example of an image with a region segmented by
the algorithm of this study.
critical, especially in applications where the conse-
quences of an incorrect prediction can be severe, such
as medical diagnosis or fraud detection.
Table 1 details the results obtained for the classi-
fication and segmentation tasks over the training and
validation. We present the loss function associated
with the classification task, allowing us to analyze
how well the model fits the training data and its poten-
tial performance on unseen data. We observe a high
initial loss in the first epoch (441.49 in training and
448.49 in validation), which is expected at the begin-
ning of the model fitting process. As the epochs pass,
the classification loss decreases dramatically, stabiliz-
ing at around 0.39 on the training set and 0.40 on the
validation set by the last epoch. This consistent re-
duction in loss suggests that the model fits the data
effectively, improving its overall performance.
Furthermore, Table 1 also reveals the values of the
specific loss function for the segmentation task. Sim-
ilar to the classification loss, the segmentation loss is
initially high (1.07 in training and 1.09 in validation
in the first epoch). However, this loss reduces signifi-
cantly over the epochs, reaching 0.22 and 0.22 in the
last epoch for the training and validation sets, respec-
tively. The drastic reduction in the segmentation loss
over the epochs indicates a substantial improvement
in the model’s ability to correctly segment the inputs,
which is crucial for applications where accurate and
detailed segmentation is required.
In Figure 5, we present a case where Gleason
Grade 3 is clearly evident. The yellow mask overlays
a region indicative of a potential cancerous area with
a Gleason Grade of 3. These preliminary results sug-
gest that there is significant potential for further refin-
ing our methodology and improving the algorithm to
achieve even greater accuracy and performance.
Comparing the results obtained with previous
studies is essential to assess the progress made and
identify areas for future improvements. Unfortu-
nately, the literature lacks studies of this nature
with the PANDA dataset. Previous studies, such
as those conducted by Silva-Rodr
´
ıguez et al. (2020)
and Arvaniti et al. (2018), have sought to classify
prostate cancer according to the Gleason scale and
have demonstrated the effectiveness of deep learning-
based approaches for the detection and classification
of prostate cancer in histopathological images. In
Silva-Rodr
´
ıguez et al. (2020), the authors developed a
proposal to assist pathologists in prostate slide analy-
sis. The work ranged from predicting Gleason grades
at the pixel level to detecting specific patterns, such
as cribriforms, to assessing the distribution of grades
in the tissue, leading to a biopsy score. The system
was based on deep learning for the Gleason Score
of prostate cancer biopsy images, using the SICAPv2
dataset, composed of 182 images, achieving an accu-
racy of 77% and reported to outperform existing state-
of-the-art methods.
In Bulten et al. (2022) authors report the results of
PANDA dataset competition. Most participants relied
on neural network architectures (such as Efficient-
Net and ResNeXt variants), different data preprocess-
ing approaches, and automated label cleaning to per-
form image classification. They also used ensembles
of multiple models, where different CNN models are
combined or the same model is trained using different
hyperparameters (such as loss function) or patch se-
lection strategies. Unfortunately, results are reported
using quadratically weighted Kappa (95% confidence
interval) on the internal validation set (0.940), which
makes it impossible to compare with ours.
Moreover, this study builds upon a strong founda-
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
704

Table 1: Training and Validation results by epoch.
Epoch Accuracy (%) False Negative Rate (%) Classification Loss Mask Loss
Training Validation Training Validation Training Validation Training Validation
1 61.39 62.36 66.47 67.52 441.49 448.49 1.07 1.09
2 83.17 82.74 98.61 98.11 37.46 37.26 0.68 0.68
3 83.63 84.96 79.79 81.06 0.72 0.74 0.46 0.47
4 86.66 88.03 68.45 69.53 0.65 0.66 0.35 0.36
5 88.72 88.12 50.17 49.84 0.68 0.67 0.34 0.33
6 89.79 88.86 34.52 34.16 0.56 0.56 0.28 0.28
7 89.47 89.78 47.59 47.75 0.62 0.62 0.28 0.28
8 89.71 89.19 36.94 36.73 0.59 0.59 0.27 0.27
9 89.79 91.09 30.28 30.71 0.52 0.53 0.26 0.26
10 90.47 90.11 30.59 30.46 0.49 0.49 0.25 0.25
11 90.43 91.62 35.70 36.17 0.49 0.50 0.25 0.26
12 90.75 90.23 26.89 26.74 0.46 0.45 0.24 0.24
13 90.50 91.30 45.74 46.14 0.54 0.55 0.29 0.29
14 90.84 90.49 25.99 25.89 0.47 0.47 0.24 0.24
15 90.95 91.30 49.10 49.28 0.61 0.61 0.29 0.29
16 91.20 91.72 29.80 29.97 0.46 0.46 0.25 0.25
17 91.15 91.13 25.23 25.23 0.45 0.45 0.23 0.23
18 91.07 90.03 29.98 29.64 0.55 0.55 0.26 0.26
19 91.25 90.57 34.02 33.77 0.49 0.48 0.25 0.25
20 91.52 91.11 37.41 37.24 0.59 0.59 0.27 0.27
21 91.52 90.84 35.31 35.05 0.49 0.49 0.26 0.26
22 91.50 91.06 25.16 25.04 0.42 0.42 0.23 0.23
23 91.97 91.34 25.36 25.19 0.43 0.43 0.23 0.23
24 91.60 91.82 22.78 22.83 0.39 0.40 0.22 0.22
25 91.03 92.23 19.59 19.84 0.36 0.37 0.19 0.20
tion by demonstrating the effectiveness of deep learn-
ing techniques in enhancing the diagnostic process of
histopathological images. The results suggest an ef-
ficient learning mechanism, capable of generalizing
well to validation data. The next steps involve exper-
imenting with different neural network architectures
and validating the model on datasets like PANDA to
further refine its performance for clinical applications.
This will enable us to expand the model’s generaliza-
tion capabilities in clinical contexts.
6 CONCLUSIONS
Prostate cancer is the most prevalent malignancy
among men, with rising rates of both incidence and
mortality worldwide. The Gleason Score, a critical
metric for assessing the histological grade of prostate
cancer, is essential in guiding therapeutic decisions
and predicting disease progression. Addressing the
growing need for efficient diagnostic tools in pub-
lic health, this study presented an approach that in-
tegrates image processing techniques and Convolu-
tional Neural Networks to analyze prostate biopsy
images, facilitating the automation of Gleason Score
segmentation and classification. The study achieved
a high accuracy of 91.34%, underscoring the po-
tential of this approach in prostate cancer diagno-
sis. Our approach enhances the precision and effi-
ciency of prostate cancer diagnostics, enabling earlier
detection and, consequently, improving patient out-
comes. Gleason Score automated analysis reduces
the reliance on subjective manual interpretation and
speeds up the diagnostic process. In future work, we
plan to evaluate other CNNs and backbones for fea-
ture extraction and to expand the datasets evaluated.
ACKNOWLEDGEMENTS
Andr
´
e R. Backes and B.A.N. Travenc¸olo grate-
fully acknowledges the financial support of CNPq
(National Council for Scientific and Technological
Development, Brazil) (Grant #307100/2021-9 and
#306436/2022-1). This study was financed in part by
the Coordenac¸
˜
ao de Aperfeic¸oamento de Pessoal de
N
´
ıvel Superior - Brazil (CAPES) - Finance Code 001.
Deep Learning for Image Analysis and Diagnosis Aid of Prostate Cancer
705

REFERENCES
Aldoj, N., Biavati, F., Michallek, F., Stober, S., and Dewey,
M. (2020). Automatic prostate and prostate zones
segmentation of magnetic resonance images using
densenet-like u-net. Scientific reports, 10(1):14315.
Arvaniti, E., Fricker, N., Moret, M., Rupp, N., Hermanns,
T., Fankhauser, C., Wey, N., Wild, P. J., Rueschoff,
J. H., and Claassen, M. (2018). Automated gleason
grading of prostate cancer tissue microarrays via deep
learning. Scientific reports, 8(1):1–11.
Brazil (2002). Programa Nacional de Controle de C
ˆ
ancer
da Pr
´
ostata: Documento de Consenso. INCA,
Bras
´
ılia, 1 edition. 1ª ed.
Bulten, W., Kartasalo, K., Chen, P., et al. (2022). Arti-
ficial intelligence for diagnosis and gleason grading
of prostate cancer: the panda challenge. Nat Med,
28:154–163. Accessed on 14/02/2024.
Chiao, J.-Y., Chen, K.-Y., Liao, K. Y.-K., Hsieh, P.-H.,
Zhang, G., and Huang, T.-C. (2019). Detection and
classification the breast tumors using mask r-cnn on
sonograms. Medicine, 98(19).
dos Santos, D. F., de Faria, P. R., Travenc¸olo, B. A., and
do Nascimento, M. Z. (2021). Automated detec-
tion of tumor regions from oral histological whole
slide images using fully convolutional neural net-
works. Biomedical Signal Processing and Control,
69:102921.
dos Santos, D. F. D., de Faria, P. R., Travenc¸olo, B. A. N.,
and do Nascimento, M. Z. (2023). Influence of data
augmentation strategies on the segmentation of oral
histological images using fully convolutional neural
networks. Journal of Digital Imaging.
Ghose, S., Oliver, A., Mart
´
ı, R., Llad
´
o, X., Vilanova, J. C.,
Freixenet, J., Mitra, J., Sidib
´
e, D., and Meriaudeau,
F. (2012). A survey of prostate segmentation method-
ologies in ultrasound, magnetic resonance and com-
puted tomography images. Computer Methods and
Programs in Biomedicine, 108(1):262–287. Accessed
on 11/02/2024.
Gonzalez, R. C. and Woods, R. E. (2008). Digital image
processing. Prentice Hall, Upper Saddle River, N.J.
Haykin, S. (1998). Neural networks: a comprehensive foun-
dation. Prentice Hall PTR.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Humphrey, P. (2004). Gleason grading and prognostic fac-
tors in carcinoma of the prostate. Mod Pathol, 17:292–
306. Published: 13 February 2004, Issue Date: 01
March 2004.
Kaggle (2023). Kaggle. https://www.kaggle.com/c/
prostate-cancer-grade-assessment. Accessed on 14
February 2024.
Kang, K. and Wang, X. (2014). Fully convolutional neu-
ral networks for crowd segmentation. arXiv preprint
arXiv:1411.4464.
Kyle, K. Y. and Hricak, H. (2000). Imaging prostate cancer.
Radiologic Clinics of North America, 38(1):59–85.
LeCun, Y., Kavukcuoglu, K., and Farabet, C. (2010). Con-
volutional networks and applications in vision. In Pro-
ceedings of 2010 IEEE International Symposium on
Circuits and Systems, pages 253–256.
Loeb, S., Bjurlin, M. A., Nicholson, J., Tammela, T. L.,
Penson, D. F., Carter, H. B., Carroll, P., and Etzioni, R.
(2014). Overdiagnosis and overtreatment of prostate
cancer. European urology, 65(6):1046–1055.
Lu, Y., Jiang, Z., Zhou, T., and Fu, S. (2019). An improved
watershed segmentation algorithm of medical tumor
image. In IOP conference series: materials science
and engineering, volume 677, page 042028. IOP Pub-
lishing.
Rodrigues, L. F., Backes, A. R., Travenc¸olo, B. A. N., and
de Oliveira, G. M. B. (2022). Optimizing a deep resid-
ual neural network with genetic algorithm for acute
lymphoblastic leukemia classification. Journal of Dig-
ital Imaging, 35(3):623–637.
Silva-Rodr
´
ıguez, J., Colomer, A., Sales, M. A., Molina, R.,
and Naranjo, V. (2020). Going deeper through the
gleason scoring scale: An automatic end-to-end sys-
tem for histology prostate grading and cribriform pat-
tern detection. Computer methods and programs in
biomedicine, 195:105637.
Society, A. C. (2023). Facts & figures 2023.
https://www.cancer.org/cancer/prostate-cancer/
about/key-statistics.html. Accessed: [02 November
2021].
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wojna,
Z. (2016). Rethinking the inception architecture for
computer vision. In IEEE Conference on Computer
Vision and Pattern Recognition (CVPR), pages 2818–
2826.
Tian, Z., Liu, L., and Fei, B. (2015). A fully automatic
multi-atlas based segmentation method for prostate mr
images. In Medical Imaging 2015: Image Processing,
volume 9413, pages 1067–1073. SPIE.
Toth, R. J., Shih, N., Tomaszewski, J. E., Feldman, M. D.,
Kutter, O., Daphne, N. Y., Paulus Jr, J. C., Pala-
dini, G., and Madabhushi, A. (2014). Histostitcher™:
An informatics software platform for reconstructing
whole-mount prostate histology using the extensible
imaging platform framework. Journal of Pathology
Informatics, 5(1):8.
Tsung-Yi Lin, M. (2015). Microsoft coco: Common objects
in context. Computer Vision and Pattern Recognition
(cs. CV), 1405.
Wu, Y. et al. (2021). Github. https://detectron2.readthedocs.
io/en/latest/. Accessed on 23 November 2023.
Wu, Y., Kirillov, A., Massa, F., Lo, W.-Y., and Girshick, R.
(2019). Detectron2: A pytorch-based modular object
detection library. arXiv preprint arXiv:1904.04514.
Yan, K., Li, C., Wang, X., Li, A., Yuan, Y., Feng, D.,
Khadra, M., and Kim, J. (2016). Automatic prostate
segmentation on mr images with deep network and
graph model. In 2016 38th Annual international con-
ference of the IEEE engineering in medicine and biol-
ogy society (EMBC), pages 635–638. IEEE.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
706
