
Semi-Supervised Anomaly Detection in Skin Lesion Images
Alina Burgert
1
, Babette Dellen
1
, Uwe Jaekel
1 a
and Dietrich Paulus
2 b
1
Faculty of Mathematics, Informatics, and Technology, University of Applied Sciences Koblenz, Joseph-Rovan-Allee 2,
53424 Remagen, Germany
2
Institute for Computational Visualistics, University Koblenz, Universit
¨
atsstraße 1, 56070 Koblenz, Germany
{burgert, dellen, jaekel}@hs-koblenz.de, paulus@uni-koblenz.de
Keywords:
Anomaly Detection, Semi-Supervised Learning, Dermatology.
Abstract:
Semi-supervised anomaly detection is the task of learning the pattern of normal samples and identifying devi-
ations from this pattern as anomalies. This approach is especially helpful in the medical domain, since healthy
samples are usually easy to collect and time-intensive annotation of training data is not necessary. In derma-
tology the utilization of this approach is not fully explored yet, since most work is limited to cancer detection,
with the normal samples being nevi. This study, instead, investigates the use of semi-supervised anomaly
detection methods for skin disease detection and localization. Due to the absence of a benchmark dataset a
custom dataset was created. Based on this dataset two different models, SimpleNet and an autoencoder, were
trained on healthy skin images only. Our experiment shows that both models are able to distinguish between
normal and abnormal samples of the test dataset, with SimpleNet achieving an AUROC score of 97 % and
the autoencoder a score of 93 %, demonstrating the potential of anomaly detection for dermatological appli-
cations. A visual analysis of corresponding anomaly maps revealed that both models have their own strengths
and weaknesses when localizing the abnormal regions.
1 INTRODUCTION
Over the last decade, deep learning methods have rev-
olutionized diagnostic capabilities in various medi-
cal domains, including dermatology. According to
Chan et al. (2020), the most common machine learn-
ing paradigm used in dermatology is supervised learn-
ing. However, supervised learning requires large an-
notated training datasets. The annotation procedure is
time-intensive and requires the expertise of medical
professionals and can introduce human bias.
An alternative approach, which addresses some of
these drawbacks, is semi-supervised anomaly detec-
tion, as defined by Chandola et al. (2009). Trans-
ferred to the medical domain, the basic idea is learn-
ing the appearance of a healthy state in order to be
able to identify pathologic cases as deviations from
this state. During training, only images showing the
healthy state are required. This is particularly useful
in cases where no or little pathological data is avail-
able and unknown pathologies also need to be recog-
nized (e. g. in the case of rare diseases). In contrast
to supervised learning, no ground truth is required for
a
https://orcid.org/0000-0002-4275-1430
b
https://orcid.org/0000-0002-2967-5277
training, which saves valuable time of medical ex-
perts. A limitation of the approach is that it does not
yield a specific diagnosis.
In medical imaging, anomaly detection is applied
predominantly to brain MRIs (Tschuchnig and Gader-
mayr, 2022). Only a few works on semi-supervised
anomaly detection in dermatology exist. Most of
them aim to detect skin cancer by learning what
normal pigmented skin lesions (nevi) look like (Lu
and Xu, 2018; Zhang et al., 2022; Grignaffini et al.,
2023; Cai et al., 2024). In contrast, other studies
learn the appearance of healthy skin without any le-
sions. However, these studies are limited to the detec-
tion of pigmented skin lesions in dermoscopic images
(Shen et al., 2020) and the detection of hand ekzema
(Gonzalez-Jimenez et al., 2023).
The aim of this work is to explore the approach of
semi-supervised anomaly detection for general skin
lesion detection and localization. Due to a lack of
benchmark datasets, we create our own skin anomaly
detection dataset. Based on this dataset, we com-
pare two anomaly detection methods, SimpleNet (Liu
et al., 2023) and a convolutional autoencoder, which
is often used as a baseline method in anomaly detec-
tion.
Burgert, A., Dellen, B., Jaekel, U. and Paulus, D.
Semi-Supervised Anomaly Detection in Skin Lesion Images.
DOI: 10.5220/0013305400003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 2: VISAPP, pages
535-541
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
535

2 STATE OF THE ART
According to Cai et al. (2024) anomaly detection
methods can be categorized into methods based on
reconstruction, self-supervised learning, and feature
reference. Reconstruction-based methods rely on
generative models, e.g., autoencoder, variational au-
toencoder, generative adversarial networks or diffu-
sion models, that are trained to reconstruct healthy
images. When reconstructing abnormal images, it is
assumed that a comparatively large reconstruction er-
ror occurs, which can be interpreted as an anomaly
score. In self-supervised learning, models are trained
on pretext tasks with generated pseudo labels. The
basic idea is that knowledge which is obtained in the
pretext task can be transferred to the anomaly detec-
tion task. Feature-reference-based methods are based
on the disparity between current and reference fea-
tures. For example, a pretrained network can be uti-
lized to extract and save features of normal images in
a memory bank for reference. During inference, fea-
tures of interest are compared to reference features in
order to detect anomalies.
Anomaly detection has also been applied in der-
matology. Lu and Xu (2018), Zhang et al. (2022),
Grignaffini et al. (2023) and Cai et al. (2024) utilize
anomaly detection for melanoma detection in dermo-
scopic images, with the normal condition being de-
fined as nevi. For example in Lu and Xu (2018), a
VAE is trained on images of nevi from the ISIC 2018
dataset. Skin diseases such as melanoma or actinic
keratosis are recognized as an anomaly with an AU-
ROC of 0.779 using a reconstruction-based approach.
In contrast to the studies above, the following
works try to detect skin lesions by learning the healthy
appearance of skin without any lesions. Shen et al.
(2020) propose a new method called adGAN for
anomaly detection. In contrast to existing GAN-based
methods, adGAN does not rely on a reconstruction er-
ror for anomaly detection. Instead, the authors follow
a discriminative approach, where fake images gen-
erated from a GAN are used as an abnormal class
and a discriminator model is trained to discriminate
between the normal and the generated abnormal im-
ages. The proposed model is tested on three datasets
including ISIC 2016 to evaluate the performance of
the model in skin lesion detection, where it achieves
an AUC value of 0.98. In Gonzalez-Jimenez et al.
(2023), a score-based diffusion model is used to de-
tect and localize hand eczema. For this purpose,
the diffusion model is trained with images of healthy
hands. The log-likelihood gradient map, which is
analysed at the beginning of the diffusion process,
is used to detect anomalies. At inference time, it is
Table 1: Number of normal and anomalous images by
source dataset used in this study.
Source # Normal # Abnormal
ISIC Archive 160 11
SD-198 11 158
ArsenicSkinImagesBD 175 -
Google Image Search - 21
All 346 190
therefore not necessary to run through the entire time-
consuming and computationally expensive diffusion
process. A test on a private dataset from a university
hospital demonstrates that hand eczema is recognized
with an AUROC of 0.912.
3 METHODS
3.1 Dataset
Since no publicly available dermatological dataset
was suitable for our anomaly detection study, we cre-
ated a custom dataset. This process involved collect-
ing two types of image classes: normal images show-
ing healthy skin to allow the model to learn the ap-
pearance of healthy skin, and abnormal images show-
ing different types of skin pathologies or irregulari-
ties to evaluate the model’s ability to detect anoma-
lies. For a skin image to be classified as healthy,
it had to show no lesions, erythema or other visi-
ble pathological signs. An exception was made for
pigmented skin lesions, since the study’s focus is
not on skin cancer detection. Pathological images
were selected to represent a variety of anomaly types
including for example erythema, psoriasis, eczema,
hematoma, scars and imprints of clothing. Only stan-
dard clinical photographs were included, while mi-
croscopic and dermoscopic images were excluded to
maintain consistency, because it could be more chal-
lenging for the model to learn generalized patterns
and appearances of skin across different zoom lev-
els. Images of certain body regions, such as hands,
feet, face, and head, were excluded because of their
unique anatomical features and variability in appear-
ance. To further simplify the task and avoid misclas-
sification of background pixels as anomalies, images
were cropped to exclude non-skin areas. Included im-
ages vary across different factors such as lightning,
skin tone, age, presence of skin folds and body hair
etc. The final dataset was created by collecting im-
ages from the following publicly available sources:
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
536

ISIC Archive
1
: The ISIC Archive, hosted by the In-
ternational Skin Imaging Collaboration (ISIC), con-
tains a large publicly available collection of skin im-
ages. The majority are dermoscopic images of pig-
mented skin lesions, which are not suitable for our
dataset. Instead we filtered the archive images for to-
tal body photographs (TBPs), which yielded 36 im-
ages, showing the posterior torso. Based on these im-
ages, we extracted multiple smaller images of compa-
rable sizes. This resulted in 160 normal images and 11
abnormal images showing scars or imprints of cloth-
ing.
ArsenicSkinImagesBD
2
: The ArsenicSkinIm-
agesBD dataset (Emu et al., 2024) contains 741
images of 37 arsenic-affected and 741 images of 76
non-arsenic-affected individuals from Bangladesh,
captured by smartphone cameras. Of the 741 non-
affected images, 175 were used as normal images.
The remaining images were excluded due to different
reasons (e.g. duplicates, showing hands / fingers or
potential skin conditions).
SD-198
3
: SD-198 (Sun et al., 2016) is a benchmark
dataset for clinical skin diseases containing 6,584 im-
ages from 198 classes. We selected 158 images from
the classes acne vulgaris, allergic contact dermatitis,
eczema, erythema annulare centrifugum, erythema
multiforme, factitial dermatitis, guttate psoriaris, pso-
riaris, tinea corporis and used them as abnormal im-
ages. In addition 11 healthy skin patches were ex-
tracted and added to the normal image dataset.
Google Image Search: Another 21 images contain-
ing erythema or hematoma were collected using a
Google Image Search and added to the abnormal
dataset.
Table 1 shows the number of normal and abnormal
images by source dataset. In total 346 normal and 190
anomalous images were collected. The dataset was
splitted into three datasets for training, validation and
evaluation. Models were trained on 250 normal im-
ages. A validation set of 62 images (20 normal and
42 abnormal) was utilized to optimize hyperparame-
ters and to save the best model for evaluation. The
test set for final evaluation contains 224 images (76
normal and 148 abnormal).
For the autoencoder, all images were resized to
128 × 128 and pixel values were scaled into a range
of [0, 1]. For SimpleNet, all images were resized to
224 × 224 and pixel values were first scaled into a
range of [0, 1] and then normalized according to the
mean and standard deviation of ImageNet as in Liu
1
https://www.isic-archive.com/
2
https://data.mendeley.com/datasets/x4hgnjj5gv/2
3
https://huggingface.co/datasets/
resyhgerwshshgdfghsdfgh/SD-198
et al. (2023). No data augmentation was applied.
3.2 Model Architectures, Training and
Evaluation
In the following we describe the two anomaly detec-
tion models, the training procedure and the evaluation
metrics used in this study.
SimpleNet: SimpleNet was introduced by Liu et al.
(2023) for the task of detecting and localizing anoma-
lies in industrial images. The authors argue that ex-
isting approaches (e. g. reconstruction- and feature-
based) have some drawbacks and therefore proposed
SimpleNet which combines several approaches and
comes with further improvements. SimpleNet con-
sists of four components. The first component is the
feature extractor, a pretrained neural network used for
extracting local image features. Since pretrained net-
works are usually trained on natural images such as
ImageNet and not on industrial or medical images, a
simple neural network called feature adaptor is uti-
lized to map the extracted features into the target do-
main. The third component is an anomalous feature
generator which artificially generates anomalous fea-
tures by adding random gaussian noise to normal fea-
tures. Last, a simple discriminator network is trained
to discriminate the normal and the artificially gener-
ated anomalous features. In contrast to Shen et al.
(2020) the discrimination is performed on individ-
ual local feature vectors, not on whole images. Sim-
pleNet with all its components can be trained in an
end-to-end fashion. During inference the generation
of anomalous features is omitted. Local features are
extracted and adapted from the input image and then
mapped to an anomaly score by the discriminator net-
work. Arranging all local anomaly scores in a 2D-
grid yields an anomaly map, highlighting anomalous
areas in the input image. Based on the anomaly map
an image level anomaly score can be computed. In
the original publication of SimpleNet the maximum
anomaly score is used.
For our experiment we used the same hyperparam-
eter configuration as in Liu et al. (2023). We trained
SimpleNet for 160 epochs with a batchsize of 8 and
saved the best model based on validation anomaly de-
tection performance.
Autoencoder: As a baseline model, we implemented
a convolutional autoencoder (AE), consisting of an
encoder and a symmetrical decoder. The encoder
compresses an input image x ∈ R
H×W ×C
into a latent
feature vector z ∈ R
d
. Based on this feature vector, the
decoder reconstructs the original image. The encoder
consists of four convolutional layers, each downsam-
pling the image resolution to
H
in
2
×
W
in
2
. The first con-
Semi-Supervised Anomaly Detection in Skin Lesion Images
537

Figure 1: Visualization of the compared methods on randomly drawn abnormal test images. The figure shows the input
abnormal images (1st row), the reconstructed images and corresponding anomaly maps generated by the autoencoder (2nd
and 3rd row) and the anomaly maps generated by SimpleNet (4th row). Warmer colors of the anomaly maps correspond to
higher pixel-level anomaly scores. Color values of anomaly maps generated by one specific model are directly comparable,
since they follow the same color scale. For visualization purposes, anomaly score outliers were cut off.
Table 2: Performance of autoencoder with different hyper-
parameter configurations. The best configuration is high-
lighted in bold.
C
0
d AUROC % AUPRC %
16 16 90.6 95.4
16 32 90.1 95.0
16 64 90.2 95.6
32 16 90.1 95.7
32 32 90.0 95.5
32 64 91.3 95.7
volutional layer has a width of C
0
channels. Each
subsequent layer increases the width by a factor of 2.
Convolutional layers are followed by a ReLU activa-
tion. The output of the last convolutional layer is flat-
tened and processed by a fully-connected layer, which
returns the feature vector of length d. A sigmoid func-
tion is used as a last activation in the decoder to ensure
that the output remains in the range of [0, 1]. Basic
width C
0
and latent dimension d were configured in
a hyperparameter optimization step by choosing the
model with the best anomaly detection performance
on the validation set (see table 2). As a reconstruction
loss, we used MSE. The model was optimized with
ADAM configured with an initial learning rate of 1e-
3, a weight decay of 1e-5 and trained for 200 epochs
with a batchsize of 8. The best model (measured in
terms of anomaly detection performance on the vali-
dation set) was updated every epoch and saved after
training.
Evaluation Metrics: Both models yield a real-
Figure 2: Receiver Operating Characteristic Curve, Preci-
sion Recall Curve and corresponding AUC values of au-
toencoder (AE) and SimpleNet (SN).
valued output which can be interpreted as an
anomaly score. Based on this score, we generated
Receiver-Operating-Characteristic- and Precision-
Recall-Curves (ROC and PRC) and calculated the
area under both curves (AUC) to evaluate the capa-
bility of the models to differentiate between normal
and abnormal images. An advantage of these met-
rics is that they do not require an additional validation
dataset for the purpose of finding an optimal decision
threshold.
4 RESULTS
The quantitative results of the models trained on
healthy skin images for the task of skin lesion de-
tection are visualized in Figure 2. The ROC-Curves
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
538
(a) (b)
(c) (d)
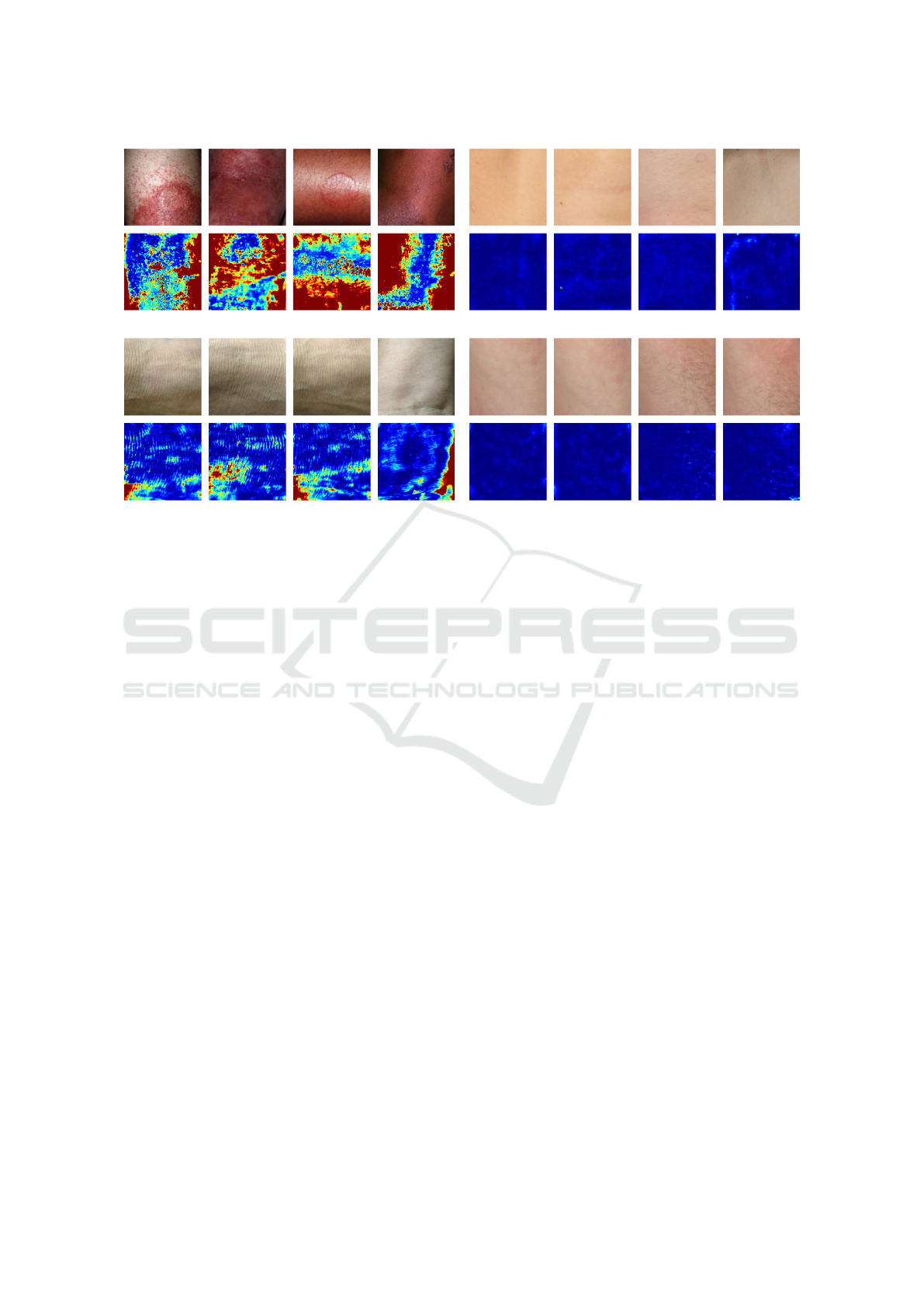
Figure 3: Visualization of (a) abnormal images with high anomaly scores, (b) abnormal images with low anomaly scores, (c)
normal images with high anomaly scores and (d) normal images with low anomaly scores based on the autoencoder.
show that both models achieve good results (Sim-
pleNet AUC 0.97, autoencoder AUC 0.93) and there-
fore are able to accurately detect skin anomalies. The
evaluation of the Precision-Recall-Curves yields sim-
ilar results (SimpleNet AUC 0.99, autoencoder AUC
0.97). In both cases, SimpleNet slightly outperforms
the autoencoder.
Furthermore, for qualitative analysis, we visual-
ize randomly drawn abnormal test images, their cor-
responding anomaly maps generated by both anomaly
detection methods and reconstruction images gener-
ated by the autoencoder in Figure 1. The anomaly
maps of the two compared models show major visual
differences. The anomaly maps generated by the au-
toencoder show fine-grained details and are, to some
extent, very good at highlighting local skin patholo-
gies. However, some anomaly maps contain large ar-
eas of false positives, often corresponding to shading
or skin folds in the input image that have not been
correctly reconstructed by the autoencoder. In con-
trast, the anomaly maps generated by SimpleNet are
less detailed, but the region containing the anomalies
is in most cases roughly highlighted.
For further qualitative analysis we sorted all nor-
mal and abnormal test images by their anomaly score
in ascending order. Four abnormal images with the
lowest and highest anomaly score as well as four
normal images with the lowest and highest anomaly
scores are visualized in Figure 3 for the autoencoder
and in Figure 4 for SimpleNet, respectively. It can be
observed that both models assign low anomaly scores
to abnormal images containing scars or imprints of
clothing (see Figure 3 (b) and 4 (b)). This is reason-
able, because these images do not contain strong con-
trasts and therefore look similar to normal images. At
the same time, normal images with strong shading e.g.
over bony prominences tend to be assigned higher
anomaly scores (see Figure 3 (c) and 4 (c)). In con-
trast, normal images with low anomaly scores look
smooth without much variation (see Figure 3 (d) and
4 (d)). SimpleNet yields particularly high anomaly
scores for abnormal images containing skin lesions
that are bright red in colour and are a strong contrast
compared to the surrounding skin (see Figure 4 (a)).
In these cases, SimpleNet is also very good at local-
izing the abnormal region which can be observed in
the corresponding anomaly map. In contrast, the au-
toencoder assigns the highest anomaly score to abnor-
mal images with very dark shadows at the image bor-
ders (see Figure 3 (a)). It appears that the reconstruc-
tion error in these regions is so large that the actual
anomaly is barely detected. This can also be observed
in some examples in Figure 1.
5 DISCUSSION AND
CONCLUSION
The aim of this study was to explore how accurately
anomaly detection methods are able to detect and lo-
Semi-Supervised Anomaly Detection in Skin Lesion Images
539
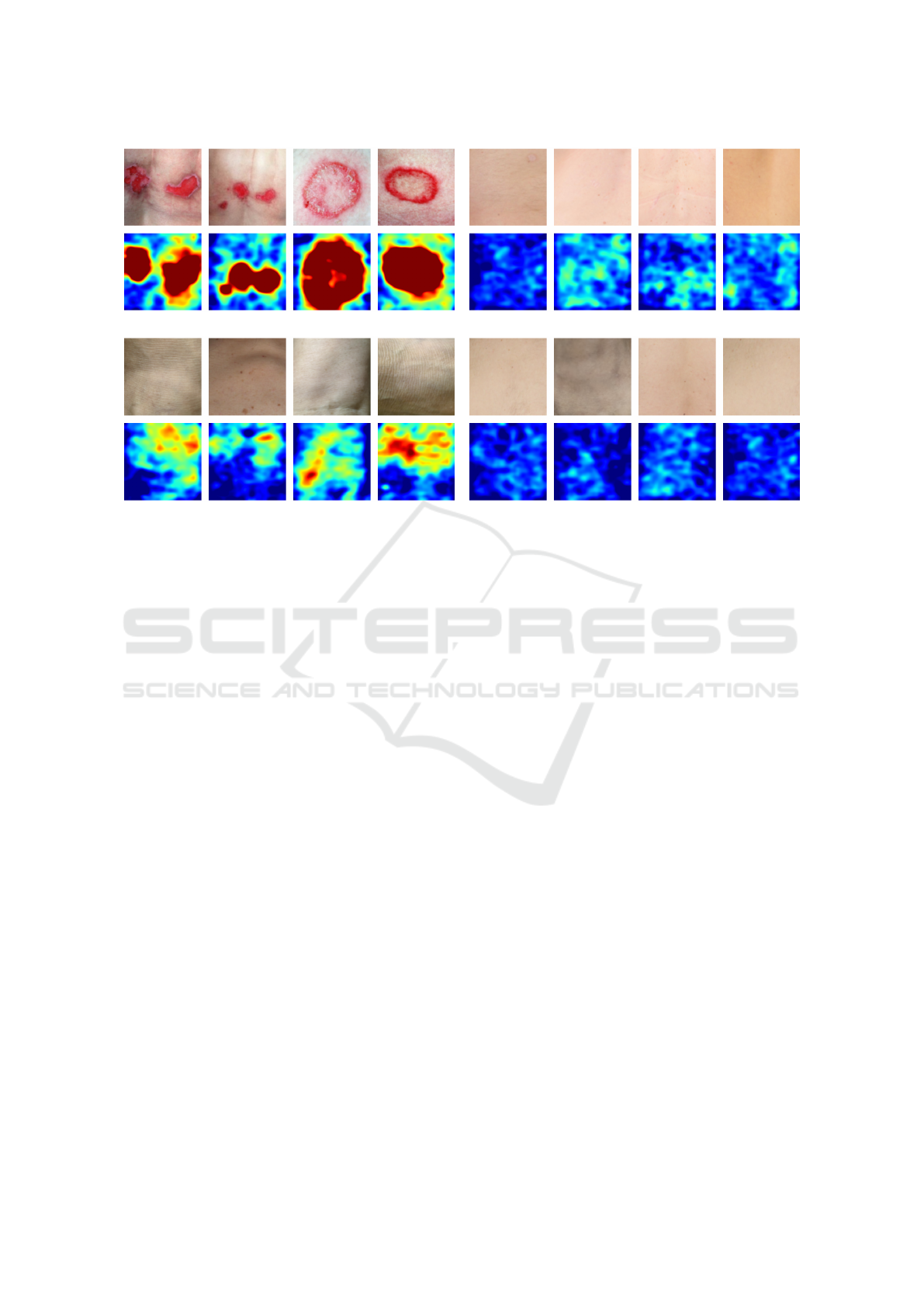
(a) (b)
(c) (d)
Figure 4: Visualization of (a) abnormal images with high anomaly scores, (b) abnormal images with low anomaly scores, (c)
normal images with high anomaly scores and (d) normal images with low anomaly scores based on SimpleNet.
calize different types of skin lesions after only being
presented images of healthy skin during training. To
answer this question a custom skin anomaly detection
dataset was created and two anomaly detection mod-
els (SimpleNet and autoencoder) were trained and
evaluated on this dataset. The results indicate that
both models are able to accurately distinguish ab-
normal images from normal images with SimpleNet
achieving an AUROC score of 97 % and the autoen-
coder a score of 93 %, respectively. Due to the ab-
sence of ground truth segmentation masks, quantita-
tive evaluation of the localization performance was
not possible. However, notable visual differences
were observed when comparing the anomaly maps
generated by each model.
Compared to SimpleNet, the autoencoder is bet-
ter at capturing fine-grained anomaly details, due to
its reconstruction-based approach. In this approach,
each anomaly score is derived from the deviation be-
tween the original and reconstructed image pixel, al-
lowing finer details to be preserved. In contrast, Sim-
pleNet calculates each anomaly score using a discrim-
inator neural network based on an image feature vec-
tor which describes the corresponding local neigh-
bourhood. As a result, details get lost during this pro-
cess.
However, the autoencoder showed high sensitiv-
ity to strong shading, frequently misclassifying it as
an abnormal region. This misclassification occurs
when shading is poorly reconstructed, resulting in
high anomaly scores that, in some cases, exceed those
of actual abnormal regions. It is possible that im-
ages with poor lighting conditions were underrepre-
sented in the training dataset, contributing to this is-
sue. In this case it would be reasonable for the model
to classify shading as abnormal. However, as long as
the overall image is correctly classified as abnormal,
pixel-level misclassifications do not impact anomaly
detection metrics like AUROC. For this reason, the
localization accuracy should be investigated quanti-
tatively in future studies to explore if the image was
classified as abnormal for the right reasons.
In addition to challenges with reconstructing
strong shading, other issues arose, such as mis-
matches in skin tone between the original and recon-
structed images. In some cases, features such as skin
folds appeared in the reconstruction even though they
were absent in the original image. Against this back-
ground, it is important to note that the autoencoder
model used for inference was selected based on the
highest validation AUROC score. Thus, the empha-
sis was on optimizing the anomaly detection perfor-
mance, rather than achieving the best possible recon-
struction quality.
A limitation of our study lies in the small sam-
ple size and diversity of our dataset, which may re-
strict the generalization ability of our model. To
further explore semi-supervised anomaly detection
in dermatology, a larger medical dataset containing
healthy skin images would be required. This data
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
540

set should exhibit a high degree of variety regard-
ing factors such as age, skin tone, presence of body
parts, body hair, and different lighting conditions. Fu-
ture work could further explore anomaly-localization
performance, which would require additional ground-
truth masks of various skin anomaly types, created by
medical experts.
ACKNOWLEDGEMENTS
This research has received funding from the Ministry
of Science and Health of Rhineland-Palatinate, Ger-
many, and the Debeka Krankenversicherungsverein
a.G. through the Forschungskolleg Data2Health.
REFERENCES
Cai, Y., Zhang, W., Chen, H., and Cheng, K.-T. (2024). Me-
dianomaly: A comparative study of anomaly detection
in medical images. arXiv preprint arXiv:2404.04518.
Chan, S., Reddy, V., Myers, B., Thibodeaux, Q., Brown-
stone, N., and Liao, W. (2020). Machine learning in
dermatology: current applications, opportunities, and
limitations. Dermatology and therapy, 10:365–386.
Chandola, V., Banerjee, A., and Kumar, V. (2009).
Anomaly detection: A survey. ACM computing sur-
veys (CSUR), 41(3):1–58.
Emu, I. A., Niloy, N. T., Karim, B. M. A., Chowdhury,
A., Johora, F. T., Hasan, M., Mittra, T., Rashid, M.
R. A., Jabid, T., Islam, M., et al. (2024). ArsenicSkin-
ImageBD: A comprehensive image dataset to classify
affected and healthy skin of arsenic-affected people.
Data in Brief, 52:110016.
Gonzalez-Jimenez, A., Lionetti, S., Pouly, M., and
Navarini, A. A. (2023). Sano: Score-based diffusion
model for anomaly localization in dermatology. In
Proceedings of the IEEE/CVF Conference on Com-
puter Vision and Pattern Recognition, pages 2988–
2994.
Grignaffini, F., Troiano, M., Barbuto, F., Simeoni, P.,
Mangini, F., D’Andrea, G., Piazzo, L., Cantisani, C.,
Musolff, N., Ricciuti, C., et al. (2023). Anomaly
detection for skin lesion images using convolutional
neural network and injection of handcrafted features:
a method that bypasses the preprocessing of dermo-
scopic images. Algorithms, 16(10):466.
Liu, Z., Zhou, Y., Xu, Y., and Wang, Z. (2023). Simplenet:
A simple network for image anomaly detection and
localization. In Proceedings of the IEEE/CVF Con-
ference on Computer Vision and Pattern Recognition,
pages 20402–20411.
Lu, Y. and Xu, P. (2018). Anomaly detection for skin
disease images using variational autoencoder. arXiv
preprint arXiv:1807.01349.
Shen, H., Chen, J., Wang, R., and Zhang, J. (2020). Coun-
terfeit anomaly using generative adversarial network
for anomaly detection. IEEE Access, 8:133051–
133062.
Sun, X., Yang, J., Sun, M., and Wang, K. (2016). A
benchmark for automatic visual classification of clini-
cal skin disease images. In Computer Vision–ECCV
2016: 14th European Conference, Amsterdam, The
Netherlands, October 11-14, 2016, Proceedings, Part
VI 14, pages 206–222. Springer.
Tschuchnig, M. E. and Gadermayr, M. (2022). Anomaly
detection in medical imaging-a mini review. In Data
Science–Analytics and Applications: Proceedings
of the 4th International Data Science Conference–
iDSC2021, pages 33–38. Springer.
Zhang, H., Guo, W., Zhang, S., Lu, H., and Zhao, X. (2022).
Unsupervised deep anomaly detection for medical im-
ages using an improved adversarial autoencoder. Jour-
nal of Digital Imaging, 35(2):153–161.
Semi-Supervised Anomaly Detection in Skin Lesion Images
541
