
Using Artificial Intelligence and Large Language Models to
Reduce the Burden of Registry Participation
James P. McGlothlin
1
and Timothy Martens
2
1
RSM US LLP, Chicago, IL, U.S.A.
2
The Heart Center, Cohen Children’s Medical Center, Queens, NY, U.S.A.
Keywords: Artificial Intelligence, Quality, Patient Safety, Registries, Analytics, Business Intelligence, OpenAI,
Large Language Models, Machine Learning, Supervised Learning.
Abstract: Health care disease registries and procedural registries serve a vital purpose in support of research and patient
quality. However, it requires a significant level of clinician effort to collect and submit the data required by
each registry. With the current shortage of qualified clinicians in the labor force, this burden is becoming even
more costly for health systems. Furthermore, the quality of the abstracted data deteriorates as over-worked
clinical staff review and abstract the data. The modern advancement in electronic medical records has actually
increased this challenge by the exponential growth in data volume per patient record. In this study, we propose
to use large language models to collect and formulate the registry data abstraction. For our initial work, we
examine popular and complicated patient registries for cardiology and cardiothoracic surgery. Initial results
demonstrate the promise of artificial intelligence and reenforce our position that this technology can be
leveraged.
1 INTRODUCTION
Patient registries are considered a vital vehicle to
enable quality and collaboration between scientists
and clinicians. Registries evaluate clinical practice,
measure patient outcomes and clinician quality and
support patient safety and research (Gliklich, 2014).
There are more than 1000 common patient registries
in use in the United States.
In an informal study at a medium-sized pediatric
hospital in the United States, we identified 29
registries in which the hospital actively participated.
The total estimated level of effort to find, collect,
input and test abstracted patient information into
these registries was estimated at over 45,000 hours a
year of clinical staff at the level of registered nurse or
higher. This included over 3,000 hours of physician
time. Clearly, the cost of collecting this data is
significant.
Despite the high cost of participation, not
participating in these registries is also not a viable
solution. Not only are the registries vital to research
and public health, but there are also financial
incentives for participation. Registries often rate
health care facilities and providers. Not only are
these rating useful for marketing purposes, they also
are often referenced by financial reimbursement
models used in value-based care and pay for
performance programs. For example, the Merit-
Based Incentive Payment System (MIPS) from the
United States Centers for Medicare & Medicaid
Services (CMS) leverages the registries used in this
project as “qualified clinical data registries” (Chen,
2017) (Blumenthal, 2017).
Large language models and generative artificial
intelligence allow textual answers to prompted
questions without training (Zhao, 2023).
Furthermore, there have been specific large language
models pre-trained on the semantics and logic innate
to medicine (Thirunavukarasu, 2023). Additionally
generative artificial intelligence can be used to search
and summarize based on specific context and
information subsets (Ghali, 2024). The authors of
this paper in previous research have had success
leveraging generative artificial intelligence for
specific health care tasks including patient chart
summarization, insurance denial appeals and clinical
trial communications. This research builds on that
success to address a larger clinical challenge.
In this position paper, we propose to utilize
generative AI in combination with advanced analytics
to populate patient registry information. Our position
McGlothlin, J. P. and Martens, T.
Using Artificial Intelligence and Large Language Models to Reduce the Burden of Registry Participation.
DOI: 10.5220/0013306200003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 789-796
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
789

is this is a good use case because it does not directly
affect acute patient care and therefore has low risk of
causing harm and because it has high potential return
on investment (ROI) due to the significant skilled
effort required to perform the task manually.
2 REGISTRY BACKGROUND
For the purpose of this experiment, we chose four
registries:
1. The Society of Thoracic Surgeons (STS)
National Database
2. The STS American College of Cardiology
(ACC) Transcatheter Valve Replacement
(TVT) Registry
3. The STS Congenital Heart Surgery Database
4. The Pediatric Cardiac Critical Care
Consortium (PC
4
)
We chose these four registries so we could limit
the experiment to a single specialty taxonomy
(cardiology and cardiothoracic surgery) and leverage
a common interface for inputting information,
without reducing our experiment to a single registry
or patient cohort. We also chose the registries due to
our previous successful experience in related research
(McGlothlin, 2018) and to the abundance of related
research.
The STS National Database has “data on nearly
10 million procedures from more than 4,300
surgeons, including 95% of adult cardiac
surgery procedures.” (http://www.sts.org/research-
data/registries/sts-national-database) (Grover, 2014).
The STS series of databases have a long and proven
history of advancing research and patient safety
(Jacobs, 2015) and the STS databases are used to
benchmark clinical outcomes and evaluate heath risks
(Wyse, 2002) (Falcoz, 2007).
Artificial intelligence including machine learning
and data mining has long been used to leverage the
STS data (Orfanoudaki, 2022) (Gandhi, 2015) (Kilic,
2020) (Scahill, 2022) for quality improvement.
However, we could not locate any significant
research leveraging AI to populate the data base in the
first place.
The STS/ACC TVT Registry includes very
specific data to study how transcatheter valve
replacement and repair procedures are being utilized.
Over 300,000 patients are in the registry and
outcomes (length of stay (LOS), mortality,
readmissions and complications) have improved
every year (Holmes, 2015) (Sorajja, 2017) (Carroll,
2020).
The STS Congenital Heart Surgery Database
contans over 600,000 congenital heart surgery
procedure records and 1,000 surgeons. In
(McGlothlin, 2018), 119 CHD diagnosis categories
were identified and data mining was able to correctly
label 78% of cases. Studies have shown that the STS
data is 80-85% accurate.
The Pediatric Cardiac Critical Care Consortium
(PC4) has detailed information on pediatric patients
in the cardiothoracic intensive care unit (CTICU).
The data has been shown to be very reliable at >99%
accurate (Gaies, 2016). In a previous experiment we
attempted to programmatically populate each data in
the PC
4
dataset. We spent 3,500 hours of
development on this project and were able to populate
over 75% of the data fields. One of the desired
outcomes of this research is to not only reduce the
clinical burden of abstraction and registry
participation but also the technical burden of
developing and maintaining custom rules for registry
population.
These registries have complex data requirements.
The STS General Thoracic Data Specifications
v5.21.1 has 215 pages describing the requirements for
data entry. The Data Dictionary Codebook
(https://med.stanford.edu/content/dam/sm/cvdi/docu
ments/pdf/sts-adult-cardiac-registry-redcap.pdf )
from Stanford University identifies 1757 data fields.
This challenge is therefore for both valuable and
sufficiently complex.
3 ACCESSING PATIENT
RECORDS
The goal of this research is to generate the precise
data fields required to enter patient records into the
registries. Thus, one of the initial requirements is we
make our AI solution have access to the needed
patient information.
To do this in a standardized way, we harness many
standards. The Fast Health Interoperability
Resources (FHIR) Standard specifies the format for
restful web APIs to communicate health care
information (Ayaz, 2021). FHIR is a standard for
health care data exchange, published by the standards
organization “HL7”. Virtually all electronic medical
record (EMR) vendors support FHIR.
For our purpose, we primarily leverage the US
core FHIR profiles (https://hl7.org/fhir/us/core/).
These specifications include allergies, care plans,
implants, diagnoses, encounters, goals,
immunizations, medications, observations, vital
HEALTHINF 2025 - 18th International Conference on Health Informatics
790

signs, interventions, patients, procedures and
specimens. Most of the data points required for the
registries is available in FHIR.
In addition to the discrete data points available
through the FHIR interface, we want to support
abstracting data from the physician notes. We pull all
notes from EMR and the details for the provider
inputting the notes. Generative artificial intelligence
performs very well with text information, so the notes
will be a primary driver in the data field population.
In previous initiatives, we have used generative AI to
process provider notes and user acceptance testing
supported our assertion that this analysis was accurate
and useful.
4 ARTIFICIAL INTELLIGENCE
As stated, the goal of this research is to use artificial
intelligence to determine the data fields to input into
each registry. For our assessment, we examine three
approaches:
• Using generative AI to populate all fields
• Using traditional AI methods, such as
machine learning and data mining, to
populate all fields
• Using a hybrid approach
Generative artificial intelligence (AI) refers to a
subset of AI models designed to create new content,
such as text, images, or data, based on patterns
learned from existing information. Unlike traditional
AI systems that classify or predict data, generative AI
uses advanced techniques like neural networks to
produce original outputs. One prominent example is
the Generative Pre-trained Transformer (GPT), which
generates human-like text by predicting the next word
in a sequence. Other types of generative AI include
image synthesis models, which can create new
images based on descriptions or input data. These
models leverage vast amounts of data to "understand"
underlying structures and generate new examples that
fit those patterns. (Fui-Hoon Nah, 2023) (Euchner,
2023) (Lv, 2023)
In healthcare, generative AI is being explored for
a variety of applications that aim to enhance
diagnostics, treatment planning, and medical
research. For instance, AI can help in generating
synthetic medical images, such as CT scans or MRIs,
to augment training datasets for radiologists or to
create realistic simulations for surgery preparation.
Additionally, generative models are used to develop
new drug compounds by predicting molecular
structures that may have therapeutic potential. AI-
driven systems also assist in personalized medicine,
creating treatment plans based on individual patient
data by analyzing patterns in medical histories,
genetic information, and other factors. With its ability
to create new insights and automate complex
processes, generative AI holds great promise in
revolutionizing healthcare by improving accuracy,
efficiency, and accessibility (Zhang, 2023) (
Shokrollahi. 2023).
For traditional artificial intelligence, we leveraged
machine learning and supervised learning. Machine
learning (ML) is a subset of artificial intelligence that
enables computers to learn from data and improve
their performance over time without being explicitly
programmed. By using algorithms that identify
patterns in large datasets, machine learning can make
predictions, classify information, and automate
decision-making processes. Techniques such as
supervised learning, where the model is trained on
labeled data, and unsupervised learning, where
patterns are identified from unlabeled data, are
commonly applied (Alpaydin, 2021). In healthcare,
ML is being used to analyze vast amounts of clinical
data, enabling healthcare professionals to make more
informed decisions. ML models are trained to
recognize patterns in patient records, medical
imaging, and genomics, improving diagnostic
accuracy and treatment recommendations(Alanazi,
2022).
In the healthcare sector, machine learning has a
wide range of applications, from early disease
detection to personalized treatment plans. ML
algorithms are used to analyze medical images for
early signs of diseases such as cancer, enabling
radiologists to identify abnormalities more efficiently
than traditional methods. In genomics, ML helps in
identifying genetic mutations that may lead to
diseases, assisting in personalized medicine
approaches. Additionally, ML is employed in
predictive analytics to forecast patient outcomes,
manage hospital resources, and predict disease
progression, improving both patient care and
operational efficiency. As healthcare systems
increasingly generate large amounts of data, machine
learning is becoming an indispensable tool in
enhancing clinical decision-making, reducing errors,
and optimizing treatment processes (Esteva, 2019;
Topol, 2019).
Supervised learning is a type of machine learning
where the model is trained on labeled data, meaning
each input is paired with the correct output. The goal
is to learn a mapping from inputs to outputs so that,
when presented with new, unseen data, the model can
predict the correct result. The process involves using
Using Artificial Intelligence and Large Language Models to Reduce the Burden of Registry Participation
791

a dataset with known labels to train the algorithm,
which then fine-tunes itself by adjusting its internal
parameters to minimize errors between predicted and
actual outcomes. This form of learning is widely used
in tasks such as classification and regression, where
the model learns to categorize data or predict
continuous values based on historical examples.
In healthcare, supervised learning has shown
significant potential in improving diagnostic
accuracy, personalized treatment plans, and
predicting patient outcomes. For instance, machine
learning models can be trained on medical images
like MRIs or X-rays, where the labels correspond to
specific diagnoses, enabling the algorithm to assist
radiologists in detecting diseases such as cancer or
tuberculosis with high accuracy. Supervised learning
is also used in predicting patient risk factors, such as
the likelihood of developing chronic diseases like
diabetes or heart disease, based on historical health
data, lifestyle choices, and genetic factors. This
application helps healthcare professionals provide
more tailored treatments and preventative measures,
thereby improving patient care and reducing overall
healthcare costs (Razzak, 2018).
Classification in artificial intelligence refers to the
process of categorizing data into predefined classes or
labels. This is a common task in machine learning,
where algorithms are trained on labeled datasets to
recognize patterns and predict outcomes for new,
unseen data. For example, classification can be used
for spam detection in emails, medical diagnoses, or
image recognition. The most widely used
classification algorithms include decision trees,
support vector machines, and neural networks.
According to Bishop (2006), machine learning
techniques such as logistic regression and naïve
Bayes are commonly employed for classification
tasks in both supervised and unsupervised learning
scenarios. Kotsiantis (2011) highlights the
importance of feature selection and preprocessing in
improving the accuracy of classification models.
Furthermore, modern advancements in deep learning
have led to the development of convolutional neural
networks (CNNs) that significantly enhance
classification performance, particularly in image and
speech recognition tasks (LeCun, 2015).
For the machine learning and supervised learning
algorithms, we trained the system by pulling
historical patient records for the electronic medical
record and extracting the submitted registry values for
those patient encounters. As the submitted values
were already manually entered by humans and tested
(reviewed) by clinicians, this method allows
supervised learning of the classification technique.
The STS entries served as our labels.
For our hybrid approach, we first allowed
generative artificial intelligence to attempt to
populate the registry values. Then, we allowed a
human to review the recommended entries. We used
this supervised learning mechanism to predict which
registry fields require human review and will need to
be changed from the generative AI response.
5 IMPLEMENTATION
APPROACH
This project is intended to be used in a commercial
setting by hospital providers, so that they can comply
with the requirements of patient registries with less
burden to hospital staff. Therefore, we wanted to only
use commercially available and respected software
products which have been approved to handle
protected health information (PHI) under the United
States’s HIPAA (Health Insurance Portability and
Accountability Act of 1996) (Moore, 2019).
Therefore, we chose to implement our work using
software available from Microsoft including Azure,
Azure Machine Learning (AML) (Barga, 2015)
(Barnes, 2015) and OpenAI.
Azure Machine Learning is a cloud-based service
provided by Microsoft to accelerate the end-to-end
machine learning lifecycle. It offers a wide range of
tools and services for building, training, and
deploying machine learning models, making it
accessible for data scientists, developers, and
businesses. Azure Machine Learning integrates with
various popular frameworks and provides capabilities
for automated machine learning (AutoML), model
versioning, and deployment in a scalable and secure
environment. Key features include automated
hyperparameter tuning, experiment tracking, and
seamless integration with Azure's cloud infrastructure
for efficient model management. Additionally, the
platform supports collaborative development with its
integrated notebooks and provides monitoring and
management tools post-deployment. Azure Machine
Learning also enables developers to create models
using both code-first and low-code experiences,
making it suitable for users at different levels of
expertise. This versatility helps businesses accelerate
their AI initiatives while maintaining governance,
security, and scalability in production systems
(Barnes, 2015).
OpenAI, a leading artificial intelligence research
HEALTHINF 2025 - 18th International Conference on Health Informatics
792
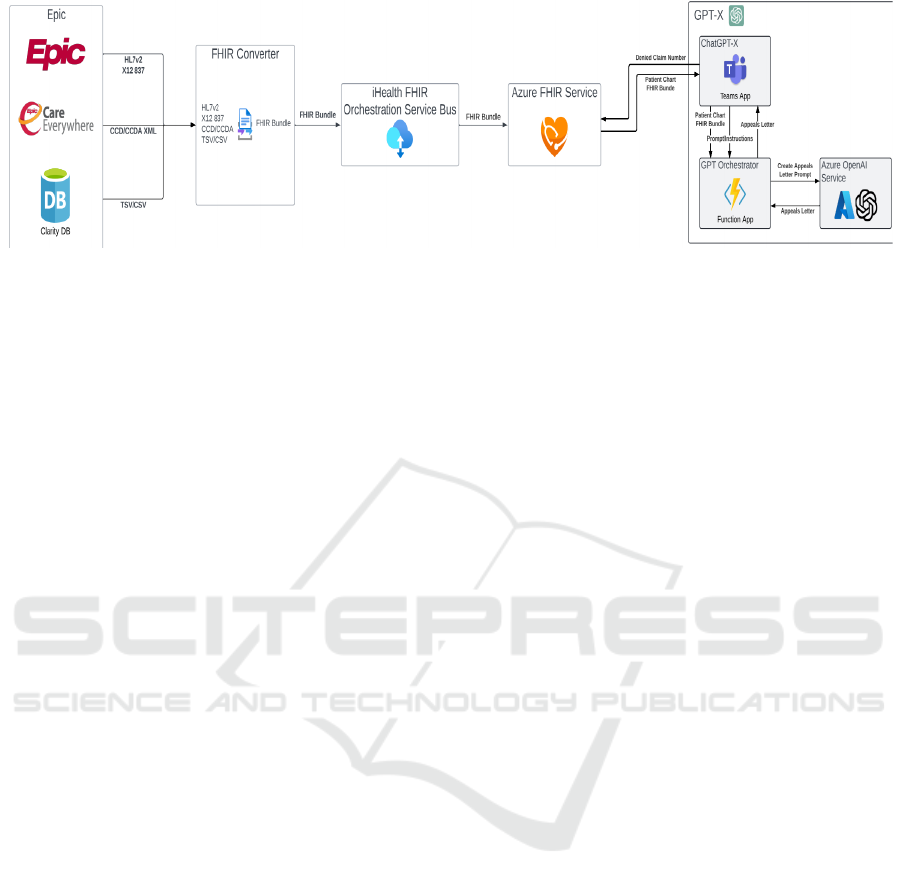
Figure 1: Architectural data flow diagram.
organization, has partnered with Microsoft to
integrate its cutting-edge AI models, like GPT, into
Microsoft Azure's cloud services. This collaboration
enables businesses and developers to leverage
powerful AI capabilities via the Azure OpenAI
Service, offering access to advanced language
models, code generation tools, and machine learning
solutions. By using Azure, users can easily scale their
AI-driven applications while benefiting from the
cloud's robust security, compliance, and flexibility.
This synergy empowers organizations to innovate
faster, automate processes, and create personalized
customer experiences while harnessing the full
potential of AI in a reliable, enterprise-grade
environment.
Microsoft Azure is enabled to support two-way
FHIR messaging. This accelerated our ability to
extract and load patient records and client data.
Figure 1 shows the implementation of the Azure
FHIR service with OpenAI utilizing the Epic
electronic medical record.
6 RESULTS
This research is in early stages of development and
validation. In order to test both the classification
technique and the generative AI approach, we attempt
to classify patient records into the appropriate
diagnosis specified by the STS Congenital Heart
Surgery Database. This classification followed the
research of (McGlothlin, 2018). Our initial results
were that when using billing diagnosis codes and
when surgery was performed, the classification
machine learning approach chose the correct
fundamental diagnoses in 98% of cases. However,
when this data was not available or accurate, or the
patient had not been surgically repaired, the accuracy
dipped significantly. Overall the diagnosis was
correct between 78% and 84% in 5 separate studies
using both generative AI and traditional machine
learning. We were unable to conclude that one
approach was significantly more accurate that the
other, it appeared to depend largely on the input data.
However, when we used our hybrid approach, starting
with the generative AI and then indicating if human
review was needed using machine learning, we were
able to improve the accuracy to 95%. In other words,
in 95% of the cases where the machine learning
algorithm predicted the generative AI classification
was accurate, it was in fact correct.
There are over 150 separate fundamental
diagnoses in version 3.2.2 of the STS Congenital
Heart Surgery Database specification. Therefore, it is
not surprising that complete accuracy was difficult to
obtain. To test our solution further, we continued to
leverage the definitions used in the STS Congenital
Heart Surgery Database, but ones with less possible
input values. Some data fields like patient name and
demographics were simply to transpose directly from
the FHIR queries and required no complex generative
AI.
The other fields chosen were premature birth,
gender, antenatal diagnosis, race, mortality status,
chromosomal abnormalities, and syndromes. Our
generative AI approach was >98% accurate across
these data fields, except for syndromes which was
93% accurate. Generative AI in combination with
machine learning was 99% accurate.
7 CHALLENGES
Many of the registry data field definitions and list of
input values change with each version upgrade. This
makes it difficult to train on historical data. We are
concerned that as the specifications changes, our
ability to predict which columns need manual review
may deteriorate.
Using Artificial Intelligence and Large Language Models to Reduce the Burden of Registry Participation
793

The patient records are often sparse. More
concerning, often the records are self-contradictory.
This complicates our artificial intelligence and
automation approach. For now, we are utilizing a set
of rules to prioritize based on timing and source data
location (for example recent claims have a higher
confidence factor).
In retrospective analysis, we should have chosen
a single registry and set of data fields upfront. We
chose a large set of related fields under the hopes that
we could decide which types of fields and patient
records the technology excels at, so that we could
focus additional phases of the initiative on the areas
with the greatest opportunity for success and return
on investment. We wanted to progress towards a
solution and methodology which was widely useful
across registries While this approach has merit, it has
stretched the time line we require to completely train
and test our model.
8 NEXT STEPS
The obvious next step is to continue testing and
training across the data fields. This will allow us to
improve the model and to accurately determine which
data fields can be automated. We recognize that
additional training, validation and statistical rigor is
needed to draw specific clinical conclusions.
Once our testing is deemed sufficient, we would
like to create an automated process. This would
allow our solution to actually populate the input
engine used by each registry. This would not only
reduce effort it would eliminate the risk of simple data
entry errors. Human review will still be part of the
process before the data is submitted.
To increase our confidence in the data and to
accelerate our testing, we would like to add a data
lineage component where the model can better
explain what data points it used to determine each
data field. Previous research has shown that
providing electronic phenotyping results improved
overall accuracy of manual chart review and reduces
the burden of clinical review (Kukhareva, 2016). Our
hope is analyzing the results and lineage will also
improve the ability of our hybrid model to predict
which entries require human review.
Finally, we hope that once our solution accurately
populates the patient registries, it can be used to
provide other actionable intelligence. One area that
interests us is “hospital at home”. This approach of
allowing an acute patient to be treated at their own
home has shown excellent results, especially for
cardiology patients. We are hoping out model can be
used to predict which patients are most likely to
achieve positive outcomes through this program.
9 CONCLUSIONS
There is no doubt that patient registry data collection
is a significant burden on health care providers. This
burden becomes more acute as the industry continues
to face staffing shortages and margin pressures.
Preliminary testing indicates that leveraging
FHIR, generative AI and machine learning in a hybrid
approach has the potential to automate the majority of
this data collection. While we are pleased with the
early results, we realize more model development and
training is needed to achieve significant results.
REFERENCES
Gliklich, R., Dreyer, N., Leavy M. (2014). Registries for
Evaluating Patient Outcomes: A User's Guide, Agency
for Healthcare Research and Quality (US). Rockville ,
MD, 3
rd
edition.
Developing a Registry of Patient Registries (RoPR). Project
Abstract. Agency for Healthcare Research and Quality;
(August 14, 2012). http://www.effectivehealthcare
.ahrq.gov/index.cfm/search-for-guides-reviews-and-
reports/?productid=690&pageaction=displayproduct.
Zhao, W. X., Zhou, K., Li, J., Tang, T., Wang, X., Hou, Y.,
... & Wen, J. R. (2023). A survey of large language
models. arXiv preprint arXiv:2303.18223.
Thirunavukarasu, A. J., Ting, D. S. J., Elangovan, K.,
Gutierrez, L., Tan, T. F., & Ting, D. S. W. (2023).
Large language models in medicine. Nature medicine,
29(8), 1930-1940.
Chang, Y., Wang, X., Wang, J., Wu, Y., Yang, L., Zhu, K.,
... & Xie, X. (2024). A survey on evaluation of large
language models. ACM Transactions on Intelligent
Systems and Technology, 15(3), 1-45.
Ghali, M. K., Farrag, A., Won, D., & Jin, Y. (2024).
Enhancing Knowledge Retrieval with In-Context
Learning and Semantic Search through Generative AI.
arXiv preprint arXiv:2406.09621.
McGlothlin, J. P., Crawford, E., Stojic, I., & Martens, T.
(2018). A Data Mining Tool and Process for Congenital
Heart Defect Management. In AMIA.
Grover, F. L., Shahian, D. M., Clark, R. E., & Edwards, F.
H. (2014). The STS national database. The Annals of
Thoracic Surgery, 97(1), S48-S54.
Jacobs, J. P., Shahian, D. M., Prager, R. L., Edwards, F. H.,
McDonald, D., Han, J. M., ... & Patterson, G. A. (2015).
Introduction to the STS National Database Series:
outcomes analysis, quality improvement, and patient
safety. The Annals of thoracic surgery, 100(6), 1992-
2000.
HEALTHINF 2025 - 18th International Conference on Health Informatics
794

Wyse, R. K., & Taylor, K. M. (2002, September). Using the
STS and multinational cardiac surgical databases to
establish risk-adjusted benchmarks for clinical
outcomes. In The heart surgery forum (Vol. 5, No. 3,
pp. E258-E264).
Stewart, J. M. (2016). Abstraction techniques for the STS
national database. The Journal of ExtraCorporeal
Technology, 48(4), 201-203.
Miele, F. (2024). Reframing Algorithms: STS perspectives
to Healthcare Automation. Springer Nature.
Orfanoudaki, A., Dearani, J. A., Shahian, D. M., Badhwar,
V., Fernandez, F., Habib, R., ... & Bertsimas, D. (2022).
Improving quality in cardiothoracic surgery: Exploiting
the untapped potential of machine learning. The Annals
of Thoracic Surgery, 114(6), 1995-2000.
Falcoz, P. E., Conti, M., Brouchet, L., Chocron, S.,
Puyraveau, M., Mercier, M., ... & Dahan, M. (2007).
The Thoracic Surgery Scoring System (Thoracoscore):
risk model for in-hospital death in 15,183 patients
requiring thoracic surgery. The Journal of Thoracic and
Cardiovascular Surgery, 133(2), 325-332.
Holmes, D. R., Nishimura, R. A., Grover, F. L., Brindis, R.
G., Carroll, J. D., Edwards, F. H., ... & STS/ACC TVT
Registry. (2015). Annual outcomes with transcatheter
valve therapy: from the STS/ACC TVT registry.
Journal of the American College of Cardiology, 66(25),
2813-2823.
Sorajja, P., Vemulapalli, S., Feldman, T., Mack, M.,
Holmes, D. R., Stebbins, A., ... & Ailawadi, G. (2017).
Outcomes with transcatheter mitral valve repair in the
United States: an STS/ACC TVT registry report.
Journal of the American College of Cardiology, 70(19),
2315-2327.
Carroll, J. D., Mack, M. J., Vemulapalli, S., Herrmann, H.
C., Gleason, T. G., Hanzel, G., ... & Bavaria, J. E.
(2020). STS-ACC TVT registry of transcatheter aortic
valve replacement. Journal of the American College of
Cardiology, 76(21), 2492-2516.
Gandhi, M., & Singh, S. N. (2015, February). Predictions
in heart disease using techniques of data mining.
In 2015 International conference on futuristic trends on
computational analysis and knowledge management
(ABLAZE) (pp. 520-525). IEEE.
Nelson, J. S., Jacobs, J. P., Bhamidipati, C. M., Yarboro, L.
T., Kumar, S. R., McDonald, D., ... & STS ACHD
Working Group. (2022). Assessment of current Society
of Thoracic Surgeons data elements for adults with
congenital heart disease. The Annals of Thoracic
Surgery, 114(6), 2323-2329.
Riehle‐Colarusso, T., Strickland, M. J., Reller, M. D.,
Mahle, W. T., Botto, L. D., Siffel, C., ... & Correa, A.
(2007). Improving the quality of surveillance data on
congenital heart defects in the metropolitan Atlanta
congenital defects program. Birth Defects Research
Part A: Clinical and Molecular Teratology, 79(11),
743-753.
Kilic, A., Goyal, A., Miller, J. K., Gjekmarkaj, E., Tam, W.
L., Gleason, T. G., ... & Dubrawksi, A. (2020).
Predictive utility of a machine learning algorithm in
estimating mortality risk in cardiac surgery. The Annals
of thoracic surgery, 109(6), 1811-1819.
Gaies, M., Donohue, J. E., Willis, G. M., Kennedy, A. T.,
Butcher, J., Scheurer, M. A., ... & Tabbutt, S. (2016).
Data integrity of the pediatric cardiac critical care
consortium (PC4) clinical registry. Cardiology in the
Young, 26(6), 1090-1096.
Scahill, C., Gaies, M., & Elhoff, J. (2022). Harnessing Data
to Drive Change: the Pediatric Cardiac Critical Care
Consortium (PC4) Experience. Current Treatment
Options in Pediatrics, 8(2), 49-63.
Ayaz, M., Pasha, M. F., Alzahrani, M. Y., Budiarto, R., &
Stiawan, D. (2021). The Fast Health Interoperability
Resources (FHIR) standard: systematic literature
review of implementations, applications, challenges
and opportunities. JMIR medical informatics, 9(7),
e21929.
Fui-Hoon Nah, F., Zheng, R., Cai, J., Siau, K., & Chen, L.
(2023). Generative AI and ChatGPT: Applications,
challenges, and AI-human collaboration. Journal of
Information Technology Case and Application
Research, 25(3), 277-304.
Euchner, J. (2023). Generative ai. Research-Technology
Management, 66(3), 71-74.
Lv, Z. (2023). Generative artificial intelligence in the
metaverse era. Cognitive Robotics, 3, 208-217.
Zhang, P., & Kamel Boulos, M. N. (2023). Generative AI
in medicine and healthcare: promises, opportunities and
challenges. Future Internet, 15(9), 286.\
Shokrollahi, Y., Yarmohammadtoosky, S., Nikahd, M. M.,
Dong, P., Li, X., & Gu, L. (2023). A comprehensive
review of generative AI in healthcare. arXiv preprint
arXiv:2310.00795.
Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M.,
Blau, H. M., & Thrun, S. (2017). Dermatologist-level
classification of skin cancer with deep neural networks.
Nature, 542(7639), 115-118.
Topol, E. (2019). Deep medicine: how artificial intelligence
can make healthcare human again. Hachette UK.
Alpaydin, E. (2021). Machine learning. MIT press.
Alanazi, A. (2022). Using machine learning for healthcare
challenges and opportunities. Informatics in Medicine
Unlocked, 30, 100924.
Razzak, M. I., Naz, S., & Zaib, A. (2018). Deep learning
for medical image processing: Overview, challenges
and the future. Classification in BioApps: Automation
of decision making, 323-350.
Li, J., Ma, Q., Chan, A. H., & Man, S. (2019). Health
monitoring through wearable technologies for older
adults: Smart wearables acceptance model. Applied
ergonomics, 75, 162-169.
Johnson, A. E., Pollard, T. J., & Mark, R. G. (2017,
November). Reproducibility in critical care: a mortality
prediction case study. In Machine learning for
healthcare conference (pp. 361-376). PMLR.
Shen, D., Wu, G., & Suk, H. I. (2017). Deep learning in
medical image analysis. Annual review of biomedical
engineering, 19(1), 221-248.
Using Artificial Intelligence and Large Language Models to Reduce the Burden of Registry Participation
795

Bishop, C. M., & Nasrabadi, N. M. (2006). Pattern
recognition and machine learning (Vol. 4, No. 4, p.
738). New York: Springer.
Kotsiantis, S. B., Zaharakis, I., & Pintelas, P. (2007).
Supervised machine learning: A review of
classification techniques. Emerging artificial
intelligence applications in computer
engineering, 160(1), 3-24.
LeCun, Y., Bengio, Y., & Hinton, G. (2015). Deep learning.
Nature, 521(7553), 436-444.
Moore, W., & Frye, S. (2019). Review of HIPAA, part 1:
history, protected health information, and privacy and
security rules. Journal of nuclear medicine
technology, 47(4), 269-272.
Barga, R., Fontama, V., Tok, W. H., & Cabrera-Cordon, L.
(2015). Predictive analytics with Microsoft Azure
machine learning (pp. 221-241). Berkely, CA: Apress.
Barnes, J. (2015). Azure machine learning. Microsoft
Azure Essentials. 1st ed, Microsoft.
Chen, M. M., Rosenkrantz, A. B., Nicola, G. N., Silva III,
E., McGinty, G., Manchikanti, L., & Hirsch, J. A.
(2017). The qualified clinical data registry: a pathway
to success within MACRA. AJNR: American Journal
of Neuroradiology, 38(7), 1292.
Blumenthal, S. (2017). The use of clinical registries in the
United States: a landscape survey. eGEMs, 5(1).
Kukhareva, P., Staes, C., Noonan, K. W., Mueller, H. L.,
Warner, P., Shields, D. E., ... & Kawamoto, K. (2017).
Single-reviewer electronic phenotyping validation in
operational settings: Comparison of strategies and
recommendations. Journal of biomedical
informatics, 66, 1-10.
HEALTHINF 2025 - 18th International Conference on Health Informatics
796
