
2.5D Deep Learning Model with Attention Mechanism for Pancreas
Segmentation on CT Scans
Idriss Cabrel Tsewalo Tondji
1,2 a
, Francesca Lizzi
2 b
Camilla Scapicchio
2 c
and
Alessandra Retico
2 d
1
Department of Computer Science, University of Pisa, Pisa, Italy
2
National Institute for Nuclear Physics, Pisa, Italy
idriss.tondji@phd.unipi.it, {francesca.lizzi, camilla.scapicchio, alessandra.retico}@pi.infn.it
Keywords:
Computed Tomography, Deep Learning, Pancreas Segmentation.
Abstract:
The accurate segmentation of the irregularly shaped pancreas on Computed Tomography (CT) scans, consist-
ing of 3D images, is a crucial but difficult part of the diagnostic evaluation of pancreatic cancer. Most current
deep learning (DL) methods tend to focus on the pancreas or the tumor separately. However, these methods
often struggle because the pancreas region is affected by the surrounding complex and low-contrast tissues.
This study aims to develop a DL system for pancreas segmentation to improve early detection of tumors.
Recognizing the powerful performance with computational demands of 3D models, 2D models appear to be
an alternative in terms of computation with a lightweight structure but they disregard the inter-slice correla-
tion which affects the performance. To address this, we are investigating the effect of the data preparation by
using a multi-channel input image on the pancreas segmentation model, which is referred to as 2.5D model.
Our method is developed and evaluated on a widely used public dataset, the Medical Segmentation Decathlon
(MSD) pancreas segmentation dataset. The 2.5D model demonstrates superior performance, reaching a Dice
Similarity Coefficient of 75.1%, surpassing the 2D segmentation model, while remaining computationally ef-
ficient.
1 INTRODUCTION
Pancreatic cancer is one of the most lethal malignan-
cies with an unfavorable prognosis (Liu et al., 2020)
and a five-year overall survival rate of 9% for pa-
tients regardless of the stage of the disease (Kami-
sawa et al., 2016). According to the Global Can-
cer Observatory (GLOBOCAN) 2020 statistics, pan-
creatic cancer accounted for approximately 466,003
deaths worldwide, with 54,277 fatalities reported in
the United States in the same year (Sung et al., 2021).
Pancreatic ductal adenocarcinoma (PDAC), the
most common form of pancreatic cancer, originates in
the exocrine glands and ducts of the pancreas (Luchini
et al., 2016). Despite advancements in cancer treat-
ment, the survival rate for PDAC remains very low,
primarily due to late diagnosis and a lack of effec-
tive treatment options (Kamisawa et al., 2016). More
a
https://orcid.org/0009-0002-6014-4199
b
https://orcid.org/0000-0003-0900-0421
c
https://orcid.org/0000-0001-5984-0408
d
https://orcid.org/0000-0001-5135-4472
than half of patients present with metastasis and 30%
have locally advanced disease at the time of diagno-
sis. As both the mortality and incidence rates of pan-
creatic cancer continue to rise globally, there is a crit-
ical need to improve survival outcomes through en-
hanced diagnostic and therapeutic approaches. Re-
cent studies have shown that patients diagnosed at
stage I can achieve a five-year survival rate of up
to 80% (Blackford et al., 2020). Therefore accurate
early detection is very crucial to enhance the prog-
nosis of PDAC. It has been enhanced by Computed
Tomography (CT) screening trials, significantly im-
proving survival rates.
Accurately segmenting the pancreas from abdom-
inal CT scans is vital for computer-aided diagnosis
(CAD) and various quantitative and qualitative anal-
yses. The volumetric images acquired with the CT
X-ray based imaging modality provide a clear view
of the pancreas. It is indeed the most commonly used
imaging technique for detecting pancreatic cancer in
clinical practice. However, visual reading and inspec-
tion of volumetric radiographic images such as CT
scans, is tedious, time-consuming, and can lead to di-
Tsewalo Tondji, I. C., Lizzi, F., Scapicchio, C. and Retico, A.
2.5D Deep Learning Model with Attention Mechanism for Pancreas Segmentation on CT Scans.
DOI: 10.5220/0013314500003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 669-675
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
669

verse sources of variability, intra and inter-operator
(Zhao et al., 2013). Thus, there is an urgent need for
a system that can automate the segmentation of pan-
creas and pancreatic tumor, assisting imaging physi-
cians in early screening, detection and quantitative
characterization of pancreatic tumor.
Unlike other abdominal organs like the liver (Li
et al., 2018), lungs (Zhao et al., 2021), and kidneys
(Bouteldja et al., 2021), which can be effectively seg-
mented using AI-based systems, the performance of
pancreatic segmentation is still not sufficient for clin-
ical practice and remains a persistent challenge. This
is due to several key challenges (Ghorpade et al.,
2023). The first one is the poor contrast around the
boundaries, the boundary of the pancreas cannot be
well defined due to the problem of fuzzy boundary
perturbation caused by the similar density of the pan-
creas and the surrounding tissues and its proximity to
other organs. The second one is the variable size and
shape of the pancreas. The shape, size, and location
of the pancreas vary greatly across individuals. This
makes it difficult for Artificial Intelligence (AI)-based
approaches to learn and represent its shape and loca-
tion. The last one is the small size of the pancreas in
the whole CT scan, as there is a serious imbalance be-
tween the size of the target and the background, which
leads to the overfitting problem on the background re-
gion. These variations introduce significant complex-
ity when attempting to accurately segment the pan-
creas and measure its volume on CT scans, an essen-
tial step for the timely diagnosis and treatment of pan-
creatic diseases, particularly pancreatic cancer. For
these reasons, despite its clinical significance, the re-
search studies focused on the pancreas segmentation
problem are less frequent compared to the ones fo-
cused on the segmentation of other abdominal organs.
In summary, the main contributions proposed in
this study are the following:
• We investigated the effect of the data prepara-
tion on the segmentation performance. We com-
bine consecutive input 2D slices to form a multi-
channel input.
• We developed a 2.5D segmentation model and
show its effectiveness on the Medical Segmen-
tation Decathlon (MSD) pancreas segmentation
dataset.
2 RELATED WORKS
Early approaches to pancreas segmentation from
abdominal CT scans primarily employed statistical
shape models. However, with the advent of deep
learning, Convolutional Neural Networks (CNNs)
quickly became the dominant technique for medical
image segmentation. Despite their powerful represen-
tational capabilities, CNN-based segmentation net-
works often struggle when applied to small organs
like the pancreas, particularly due to the varied back-
ground content in CT images. This inconsistency can
degrade performance and result in suboptimal seg-
mentation outcomes. To counteract these challenges,
some methods attempt to refine the region of interest
(ROIs) before performing dense predictions, yet such
approaches only partially mitigate the issue.
State-of-the-art methods in pancreas segmentation
can be divided into two main categories: 2D and 3D
segmentation networks. The selection of 2D or 3D
networks often hinges on the specific application re-
quirements and the availability of computational re-
sources. In 2D networks, the data is sliced along im-
age planes, and each slice is independently fed into
the model (Zhang et al., 2021). While this approach
is computationally efficient, it fails to capture the full
spatial context, potentially limiting segmentation ac-
curacy. This is especially problematic when analyz-
ing volumetric CT data, as 2D networks lack the abil-
ity to extract inter-slice relationships (Wang et al.,
2021). In contrast, 3D models process entire CT vol-
umes, providing a richer representation of volumet-
ric relationships but at the cost of significantly higher
computational requirements (Yan and Zhang, 2021).
U-Net, a popular neural network for biomedical
image analysis (Ronneberger et al., 2015), has been
widely adopted for pancreas segmentation on CT im-
ages (Huang et al., 2022). The symmetric encoder-
decoder structure with skip connections allows U-Net
to efficiently capture both local and global features
(Ronneberger et al., 2015). The encoder extracts key
features through convolution and pooling operations
(Litjens et al., 2017), while the decoder restores the
image through upsampling (Milletari et al., 2016).
However, the U-Net performance is often subopti-
mal when dealing with organs as small and irregu-
larly shaped as the pancreas. Ghorpade et al. (Ghor-
pade et al., ) proposed a hybrid two-stage U-Net for
segmenting both the pancreas and pancreatic tumors,
while Milletari et al. (Milletari et al., 2016) intro-
duced V-Net, a 3D counterpart of U-Net with residual
convolutional units. Although these models demon-
strate improved performance, the pancreas occupies
less than 2% of the total CT volume, and its blurred
boundaries often confuse the network, leading to in-
accurate segmentation.
With the help of an attention mechanism, the net-
work can focus on the most relevant features without
extra supervision. For example, Oktay et al. (Oktay
et al., 2018) proposed attention U-net, which can eas-
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
670

ily integrate attention gates into the U-net model with
increasing minimal computational resources while
improving the segmentation performance.
Alves et al. (Alves et al., 2022) used the nn-UNet
for the detection and segmentation of pancreas and
pancreatic tumor. The nn-UNet architecture achieved
better performance for the pancreas and showcased
better results for tumor detection. However, a small
receptive field of CNNs may limit their ability to cap-
ture distant regions and, to some extent, overlooks
valuable global context, making it challenging to fur-
ther enhance network performance.
This paper introduces a modified version of Atten-
tion U-net focusing on increasing the receptive field
of CNNs for extracting effective features. Our main
goal is to evaluate the effect of using adjacent slices
as multi-channel input compared to the use of only
one single slice for the pancreas segmentation task.
Furthermore, we present our findings from analyzing
CT images in the Medical Segmentation Decathlon
(MSD) dataset, which is described in the next section.
3 METHODS
3.1 Dataset
MSD Tumors-Pancreas Dataset (Simpson et al.,
2019): This dataset comprises 281 abdominal
contrast-enhanced CT scans in the NIfTI format and
includes labeled masks of pancreas and pancreatic tu-
mors (see Fig. 1). This dataset is sourced from the
Medical Segmentation Decathlon (MSD) pancreas
segmentation dataset. Each CT volume has a reso-
lution of 512 x 512 x L pixels, where L belonging
to [37, 751] is the number of slices along the third
axis. For our pancreas segmentation experiments,
we consider the union of the pancreas category and
the tumor category as the target category. The data
set is available here (https://drive.google.com/drive/
folders/1HqEgzS8BV2c7xYNrZdEAnrHk7osJJ–2).
Figure 1: One sample CT slice of the MSD dataset: original
image (left), and the corresponding mask (right).
3.2 Preprocessing Step
Some pre-processing operations have been applied to
improve the quality of CT scans, The first one is the
Hounsfield Units (HU) windowing which consists of
selecting a specific range of grey values to enhance
the appearance of the tissues of interest. We applied
two different windows, clipping the pixel grey values
to reside within the range of [-200, 300] or [-100, 240]
, to see whether they produce different effects.
Images are normalized according to min-max nor-
malization in the range [0, 1]. Intensity normaliza-
tion is standardizing the pixel values such that images
should have consistent pixel values for segmentation,
potentially improving the model’s consistency, train-
ing stability and performance.
Given the computational demands of 3D convolu-
tional neural networks, particularly during the initial
exploration phase, it was chosen to simplify the task
by taking 2D slices from the 3D CT volumes. We
extracted slices along the axial plane, effectively con-
verting 3D data into 2D images.
Finally, CT images are resized to a dimension of
256x256 pixels, this helps to change the resolution
or spatial dimensions of the CT scans to achieve the
desired resolution.
3.3 Modified Attention U-Net Model
The Attention U-Net is an advanced variation of the
U-Net model designed for medical image segmenta-
tion tasks, where precise localization of regions of
interest is crucial. It incorporates attention mecha-
nisms into the traditional U-Net architecture to en-
hance the model’s ability to focus on relevant regions
in the input images while suppressing irrelevant back-
ground information (Oktay et al., 2018). These atten-
tion mechanisms are introduced in the skip connec-
tions as illustrated in Fig. 2, enabling the model to
learn which spatial regions to emphasize based on the
features propagated from the encoder to the decoder.
This selective attention improves segmentation accu-
racy, especially in cases with complex or small struc-
tures.
The model works by generating attention maps
that dynamically weigh the importance of spatial fea-
tures, depending on the task at hand. This process
allows the network to filter out less relevant fea-
tures before merging the encoder and decoder paths.
By combining the U-Net’s strength in localization
with attention’s focus mechanism, the Attention U-
Net achieves better performance in segmenting chal-
lenging datasets. It is particularly effective in scenar-
ios where there is a significant imbalance between the
2.5D Deep Learning Model with Attention Mechanism for Pancreas Segmentation on CT Scans
671

size of the target structures and the surrounding con-
text.
Similar to the Attention U-Net, the modified ar-
chitecture consists of an encoder and decoder, each
with four blocks. However, unlike the original de-
sign, our modified version incorporates dense con-
volutional layers to expand the receptive field, en-
hancing the network’s ability to extract effective fea-
tures for segmenting contextual regions. By increas-
ing the effective receptive field, deeper neurons are
connected to a larger portion of the input image, en-
abling the model to capture more contextual informa-
tion. This is particularly valuable for segmentation
tasks, where global context plays a crucial role in ac-
curately identifying and delineating regions of inter-
est.
Figure 2: Overview of the Attention U-net model architec-
ture (Oktay et al., 2018).
3.4 2.5D Network
In this section, we present a novel 2.5D segmenta-
tion network designed to address the limitations of
2D and 3D segmentation models. While U-Net has
demonstrated exceptional performance in medical im-
age segmentation, traditional 3D networks require
substantial computational resources, and 2D models
struggle to capture spatial information along the third
dimension. To overcome these challenges, our 2.5D
approach combines the efficiency of 2D convolutional
layers with the ability to extract inter-slice features
by incorporating 3D spatial context. We use adja-
cent slices to form multi-channel input images, al-
lowing the network to leverage 3D information with-
out the computational complexity of a full 3D model.
This design strikes a balance between accuracy and
resource efficiency, enabling the extraction of mean-
ingful inter-slice features while requiring significantly
fewer resources than 3D networks. Specifically, We
examine the use of three (03) input slices to form n-
channel input (n=3), comprising the central slice and
one slice from each side described as (S
i−1
, S
i
, S
i+1
),
where S
i
represents the i-th slice or middle slice.
3.5 Implementation Details
We implemented a modified version of the Atten-
tion U-Net model from scratch using the TensorFlow
framework. The model is trained for 50 epochs with
an Adam optimizer. The learning rate is set to 0.0001
and batch size is set to 4. This model incorporates
denser convolutional layers and an attention mecha-
nism to help focus the network attention on the rel-
evant regions of interest, i.e. on the pancreas. Addi-
tionally, we introduced L2 regularisation during train-
ing, a technique to prevent the model’s weights from
becoming too large and potentially leading to over-
fitting. The dataset has been partitioned into training,
test and validation sets, allocating 70% (200 patients),
20% (50 patients), and 10%(32 patients) of the data
to each set, respectively. The validation set is set to
check for overfitting.
We employ a weighted combination of Binary
Cross Entropy (BCE) loss and Dice loss to effectively
balance the contributions of both losses as described
by (He et al., 2024).
4 RESULTS
4.1 Evaluation Metric
The Dice Similarity Coefficient (DSC) has been used
as the primary metric to evaluate the model perfor-
mance. The DSC measures the similarity between the
segmentation mask predicted by the model and the
ground truth annotation. The formula is defined as
follow (Xia et al., 2024):
DSC =
2|y
g
∩ y
p
|
|y
g
| + |y
p
|
;
where y
p
and y
g
represent the prediction and the
ground truth, respectively.
4.2 Evaluation and Analysis
The results of the training in terms of average Dice
score for the test subset of MSD dataset, are reported
in Table 1. Different intensity windowing strategy
have been used, [-200, 300] for model 1 and [-100,
240] for model 2. We trained a U-net model and our
revised version of Attention-Unet.
From the table 1, we can observe that not much
variation in performance has been achieved by vary-
ing the intensity windowing strategy and an improv-
ment of the performance for pancreatic segmentation
has been achieved with the 2.5D model, demonstrat-
ing the potential benefits of the 2.5D network.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
672
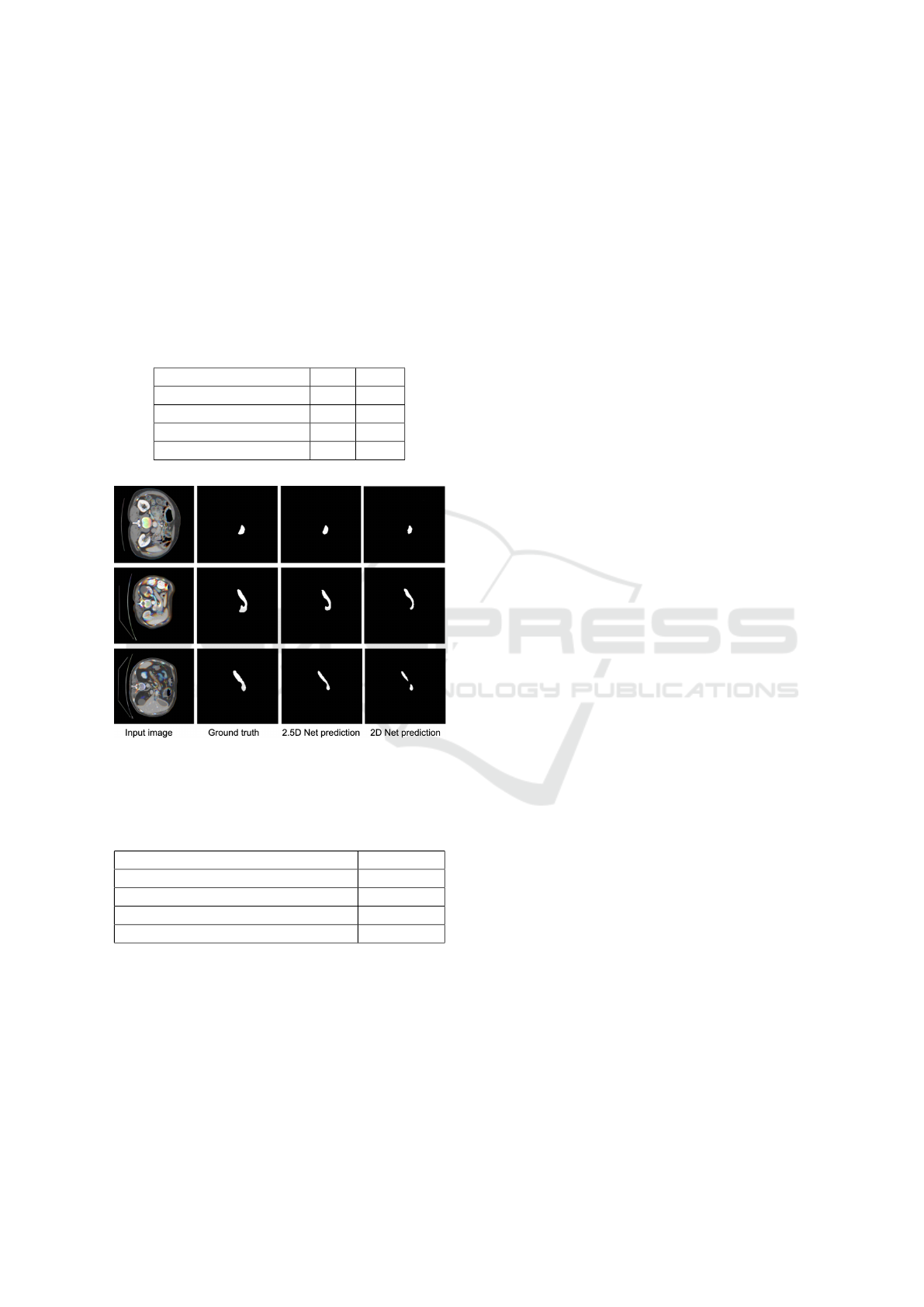
The Dice score of the model with attention mech-
anism surpasses that obtained with the model without
a attention module. This is because the attention mod-
ule helps the network to focus on the relevant regions
of interest. Three examples of segmentation obtained
on test samples of the MSD dataset is illustrated in
Fig. 3. We can clearly visualize the result performed
by the two models. The 2.5D model is able to predict
correctly the mask of the pancreas.
Table 1: Segmentation results on the test set in terms of Dice
Similarity Coefficient (DSC) on the MSD Pancreas dataset.
model 2D 2.5D
Unet1 70.1 72.2
Unet2 70.5 72.4
Modified Atten-Unet1 71.1 74.8
Modified Atten-Unet2 71.3 75.1
Figure 3: Qualitative results of three different inputs. From
left to right, we have the input image, the corresponding
mask and the prediction.
Table 2: Comparison of segmentation results in Dice score
in the MSD dataset.
Method Dice Score
(Zhu et al., 2019) 79.94
UMRFormer-Net (Fang et al., 2023) 77.36
MDAG-Net (Cao et al., 2023) 83.39
(Fang et al., 2019) 84.71
When comparing methods applied to the MSD
dataset, as shown in Table 2, we observe that our cur-
rent results exhibit a lower Dice coefficient than those
reported in the literature. This discrepancy can be ex-
plained by the fact that most state-of-the-art methods
in the literature are based on 3D models. In future
steps, we plan to transition to 3D models, which we
expect will lead to significant improvements in per-
formance.
5 CONCLUSIONS
In this paper, we develop a 2.5D model with a modi-
fied version of Attention U-Net designed for pancreas
segmentation. The model incorporates an attention
module that enables the network to focus on relevant
regions while reducing the influence of background
noise. To enhance computational efficiency, our 2.5D
U-Net relies exclusively on 2D convolutional layers
and processes 3 adjacent slices as a 3-channel input.
This approach effectively captures inter-slice infor-
mation, achieving a higher Dice coefficient compared
to 3D networks while requiring fewer computational
resources.
We evaluate our model on the MSD pancreas
dataset, demonstrating its effectiveness. Addition-
ally, the results highlight the significant impact of data
preparation on segmentation performance, underscor-
ing the importance of preprocessing in medical imag-
ing tasks.
As future work, we aim to evaluate our method
on another benchmark pancreas dataset. Additionally,
we plan to develop more advanced and robust 2.5D
models, such as transformer-based architectures, and
conduct comprehensive comparisons with state-of-
the-art models.
ACKNOWLEDGEMENTS
Research partly supported by European Commission
under the NextGeneration EU programme through
the projects: PNRR - M4C2 - I1.4, CN00000013 -
ICSC – Centro Nazionale di Ricerca in High Perfor-
mance Computing, Big Data and Quantum Comput-
ing - Spoke 8 In Silico medicine and Omics Data;
PNRR - M4C2 - I1.3, PE00000013 - FAIR - Future
Artificial Intelligence Research - Spoke 8 Pervasive
AI; PNRR - M4C2 - I1.5 - ECS00000017 Tuscany
Health Ecosystem (THE) - Spoke 1 Advanced Radio-
therapies and Diagnostics in Oncology.
REFERENCES
Alves, N., Schuurmans, M., Litjens, G., Bosma, J. S., Her-
mans, J., and Huisman, H. (2022). Fully automatic
deep learning framework for pancreatic ductal adeno-
carcinoma detection on computed tomography. Can-
cers, 14(2):376.
Blackford, A. L., Canto, M. I., Klein, A. P., Hruban, R. H.,
and Goggins, M. (2020). Recent trends in the in-
cidence and survival of stage 1a pancreatic cancer:
a surveillance, epidemiology, and end results analy-
2.5D Deep Learning Model with Attention Mechanism for Pancreas Segmentation on CT Scans
673

sis. JNCI: Journal of the National Cancer Institute,
112(11):1162–1169.
Bouteldja, N., Klinkhammer, B. M., B
¨
ulow, R. D., Droste,
P., Otten, S. W., Von Stillfried, S. F., Moellmann,
J., Sheehan, S. M., Korstanje, R., Menzel, S., et al.
(2021). Deep learning–based segmentation and quan-
tification in experimental kidney histopathology. Jour-
nal of the American Society of Nephrology, 32(1):52–
68.
Cao, L., Li, J., and Chen, S. (2023). Multi-target segmenta-
tion of pancreas and pancreatic tumor based on fusion
of attention mechanism. Biomedical Signal Process-
ing and Control, 79:104170.
Fang, C., Li, G., Pan, C., Li, Y., and Yu, Y. (2019). Glob-
ally guided progressive fusion network for 3d pan-
creas segmentation. In Medical Image Computing and
Computer Assisted Intervention–MICCAI 2019: 22nd
International Conference, Shenzhen, China, October
13–17, 2019, Proceedings, Part II 22, pages 210–218.
Springer.
Fang, K., He, B., Liu, L., Hu, H., Fang, C., Huang, X., and
Jia, F. (2023). Umrformer-net: a three-dimensional
u-shaped pancreas segmentation method based on a
double-layer bridged transformer network. Quantita-
tive Imaging in Medicine and Surgery, 13(3):1619.
Ghorpade, H., Jagtap, J., Patil, S., Kotecha, K., Abraham,
A., Horvat, N., and Chakraborty, J. (2023). Automatic
segmentation of pancreas and pancreatic tumor: A re-
view of a decade of research. IEEE Access.
Ghorpade, H., Kolhar, S., Jagtap, J., and Chakraborty, J. An
optimized two stage u-net approach for segmentation
of pancreas and pancreatic tumor. Available at SSRN
4876121.
He, J., Luo, Z., Lian, S., Su, S., and Li, S. (2024). To-
wards accurate abdominal tumor segmentation: A 2d
model with position-aware and key slice feature shar-
ing. Computers in Biology and Medicine, 179:108743.
Huang, B., Huang, H., Zhang, S., Zhang, D., Shi, Q., Liu,
J., and Guo, J. (2022). Artificial intelligence in pan-
creatic cancer. Theranostics, 12(16):6931.
Kamisawa, T., Wood, L. D., Itoi, T., and Takaori, K. (2016).
Pancreatic cancer. The Lancet, 388(10039):73–85.
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.-W., and Heng,
P.-A. (2018). H-denseunet: hybrid densely con-
nected unet for liver and tumor segmentation from
ct volumes. IEEE transactions on medical imaging,
37(12):2663–2674.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., Van Der Laak, J. A.,
Van Ginneken, B., and S
´
anchez, C. I. (2017). A survey
on deep learning in medical image analysis. Medical
image analysis, 42:60–88.
Liu, Y., Feng, M., Chen, H., Yang, G., Qiu, J., Zhao, F., Cao,
Z., Luo, W., Xiao, J., You, L., et al. (2020). Mechanis-
tic target of rapamycin in the tumor microenvironment
and its potential as a therapeutic target for pancreatic
cancer. Cancer letters, 485:1–13.
Luchini, C., Capelli, P., and Scarpa, A. (2016). Pancre-
atic ductal adenocarcinoma and its variants. Surgical
pathology clinics, 9(4):547–560.
Milletari, F., Navab, N., and Ahmadi, S.-A. (2016). V-
net: Fully convolutional neural networks for volumet-
ric medical image segmentation. In 2016 fourth inter-
national conference on 3D vision (3DV), pages 565–
571. Ieee.
Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich,
M., Misawa, K., Mori, K., McDonagh, S., Hammerla,
N. Y., Kainz, B., et al. (2018). Attention u-net: Learn-
ing where to look for the pancreas. arXiv preprint
arXiv:1804.03999.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical image computing and
computer-assisted intervention–MICCAI 2015: 18th
international conference, Munich, Germany, October
5-9, 2015, proceedings, part III 18, pages 234–241.
Springer.
Simpson, A. L., Antonelli, M., Bakas, S., Bilello, M.,
Farahani, K., van Ginneken, B., Kopp-Schneider, A.,
Landman, B. A., Litjens, G., Menze, B., Ronneberger,
O., Summers, R. M., Bilic, P., Christ, P. F., Do,
R. K. G., Gollub, M., Golia-Pernicka, J., Heckers,
S. H., Jarnagin, W. R., McHugo, M. K., Napel, S.,
Vorontsov, E., Maier-Hein, L., and Cardoso, M. J.
(2019). A large annotated medical image dataset for
the development and evaluation of segmentation algo-
rithms.
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjo-
mataram, I., Jemal, A., and Bray, F. (2021). Global
cancer statistics 2020: Globocan estimates of in-
cidence and mortality worldwide for 36 cancers in
185 countries. CA: a cancer journal for clinicians,
71(3):209–249.
Wang, Y., Zhang, J., Cui, H., Zhang, Y., and Xia, Y.
(2021). View adaptive learning for pancreas segmen-
tation. Biomedical Signal Processing and Control,
66:102347.
Xia, F., Peng, Y., Wang, J., and Chen, X. (2024). A 2.5
d multi-path fusion network framework with focus-
ing on z-axis 3d joint for medical image segmen-
tation. Biomedical Signal Processing and Control,
91:106049.
Yan, Y. and Zhang, D. (2021). Multi-scale u-like network
with attention mechanism for automatic pancreas seg-
mentation. PLoS One, 16(5):e0252287.
Zhang, Y., Wu, J., Liu, Y., Chen, Y., Chen, W., Wu, E. X.,
Li, C., and Tang, X. (2021). A deep learning frame-
work for pancreas segmentation with multi-atlas reg-
istration and 3d level-set. Medical Image Analysis,
68:101884.
Zhao, B., Tan, Y., Bell, D. J., Marley, S. E., Guo, P., Mann,
H., Scott, M. L., Schwartz, L. H., and Ghiorghiu,
D. C. (2013). Exploring intra-and inter-reader vari-
ability in uni-dimensional, bi-dimensional, and volu-
metric measurements of solid tumors on ct scans re-
constructed at different slice intervals. European jour-
nal of radiology, 82(6):959–968.
Zhao, C., Xu, Y., He, Z., Tang, J., Zhang, Y., Han, J.,
Shi, Y., and Zhou, W. (2021). Lung segmentation
and automatic detection of covid-19 using radiomic
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
674

features from chest ct images. Pattern Recognition,
119:108071.
Zhu, Z., Liu, C., Yang, D., Yuille, A., and Xu, D. (2019). V-
nas: Neural architecture search for volumetric medical
image segmentation. In 2019 International conference
on 3d vision (3DV), pages 240–248. IEEE.
2.5D Deep Learning Model with Attention Mechanism for Pancreas Segmentation on CT Scans
675
