
Challenges of Generalizing Machine Learning Models in Healthcare
Steven Kessler
a
, Bastian Dewitz
b
, Santhoshkumar Sundarara, Favio Salinas, Artur Lichtenberg
c
,
Falko Schmid
d
and Hug Aubin
e
Digital Health Lab Düsseldorf, University Clinic Düsseldorf, Moorenstrasse 5, Düsseldorf, Germany
{steven.kessler, bastian.dewitz, santhoshkumar.sundararaj, favioernesto.salinassoto, falko.schmid, hug.aubin,
Keywords:
Generalization, Machine Learning, Healthcare, Data Analysis.
Abstract:
Generalization problems are common in machine learning models, particularly in healthcare applications.
This study addresses the issue of real-world generalization and its challenges by analyzing a specific use case:
predicting patient readmissions using a Recurrent Neural Network (RNN). Although a previously developed
RNN model achieved robust results on the Medical Information Mart for Intensive Care (MIMIC-III) dataset,
it showed near-random predictive accuracy when applied to the local hospital’s data (Moazemi et al., 2022).
We hypothesize that this discrepancy is due to patient demographics, clinical practices, data collection meth-
ods, and healthcare differences in infrastructure. By employing statistical methods and distance metrics for
time series, we identified critical disparities in demographic and vital data between the MIMIC and hospital
data. These findings highlight possible challenges in developing generalizable machine learning models in
healthcare environments and the need to improve not just algorithmic solutions but also the process of mea-
suring and collecting medical data.
1 INTRODUCTION
Machine Learning, especially deep learning applica-
tions, are becoming more common in healthcare (Ku-
mari et al., 2023). The vast availability of data via
public data sets, such as MIMIC (Johnson et al., 2023)
and established electronic health record systems, al-
lows for building end-to-end models via deep learn-
ing methods. These can then be used to build de-
cision support systems (Al-Zaiti et al., 2023; Alaa
et al., 2019) or to extract knowledge (Shapiro et al.,
2023) without necessarily relying on expert knowl-
edge or detailed preprocessing. Instead of human-
engineered features and rules, deep learning models
rely on a large dataset, especially if the data is multi-
variate. They may fail to learn properly if only limited
data is available. State-of-the-art deep learning mod-
els often outperform previously established methods,
which may lead to better healthcare and patient out-
comes. However, at the current state, the dependency
on large, digitally available datasets would limit the
a
https://orcid.org/0009-0006-1770-8541
b
https://orcid.org/0000-0002-0775-1056
c
https://orcid.org/0000-0001-8580-6369
d
https://orcid.org/0009-0005-3745-2021
e
https://orcid.org/0000-0001-9289-8927
application of deep learning models to large institu-
tions and healthcare providers that treat enough pa-
tients to collect the necessary amount and type of data
(Panch et al., 2019).
A possible solution could be to develop models
that truly generalize so they can be used to make pre-
dictions and classifications on new, independent data
sets. Machine Learning models are usually evaluated
on the same dataset used for training the model, uti-
lizing a subset of the data not used for training. Even
if the model performs well across all metrics, it may
fail for similar but truly independent data.
In the study (Moazemi et al., 2022), an accurate
prediction model was trained on multivariate time se-
ries data from MIMIC-III (Johnson et al., 2023) and
evaluated on a smaller COPRA dataset
1
originating
from the patient data management system (PDMS)
of a local university hospital. The goal of the model
was to classify whether patients are likely to be read-
mitted to the ICU within a specific time frame after
discharge, making this a binary classification prob-
lem. Both datasets include patient data collected pri-
marily from the intensive care unit (ICU) and hos-
pital records, covering demographic information, vi-
tal signs, and laboratory results. The datasets were
1
This dataset is not publicly available.
254
Kessler, S., Dewitz, B., Sundarara, S., Salinas, F., Lichtenberg, A., Schmid, F. and Aubin, H.
Challenges of Generalizing Machine Learning Models in Healthcare.
DOI: 10.5220/0013320200003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 254-260
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
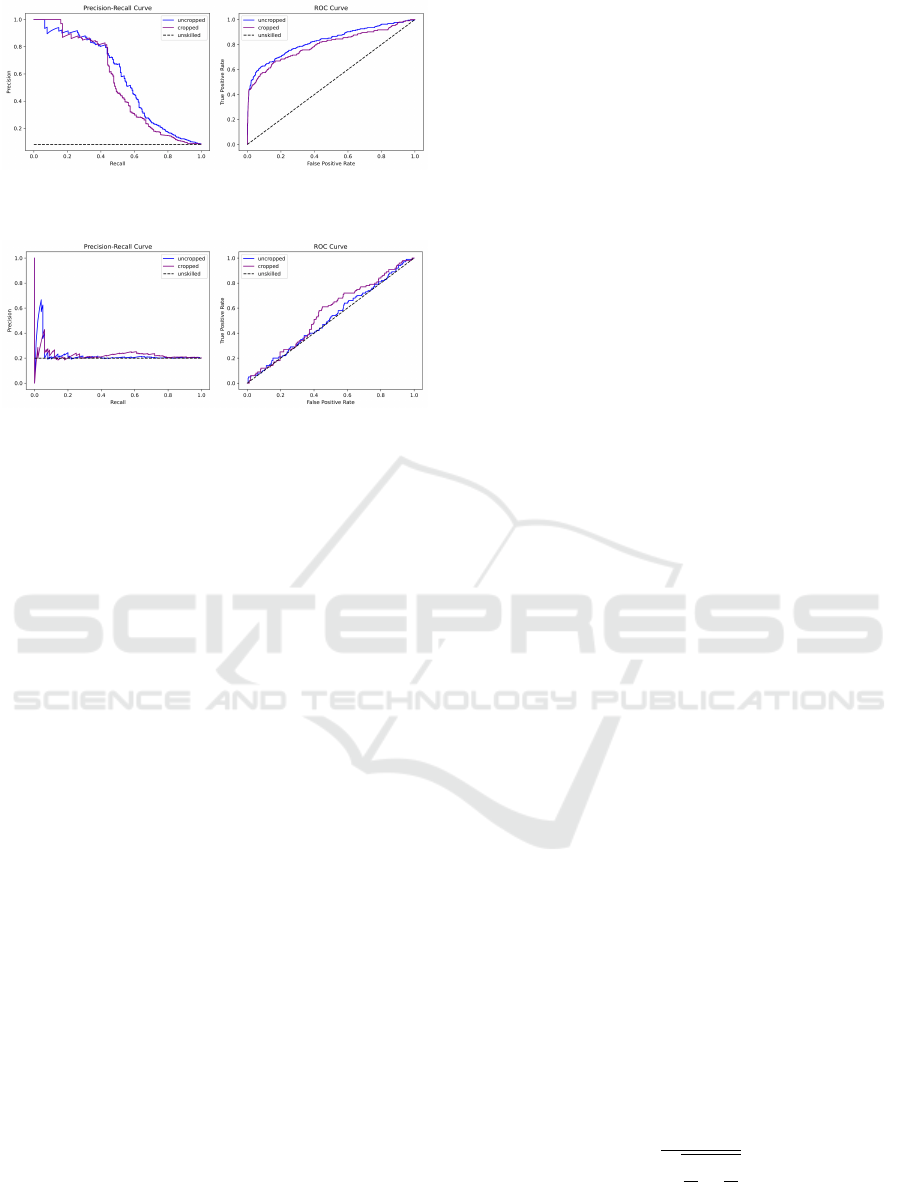
Figure 1: Evaluation results for two models trained and
evaluated on MIMIC-III. (Moazemi et al., 2022).
Figure 2: Evaluation results for two models trained on
MIMIC-III and evaluated on COPRA(Moazemi et al.,
2022).
prepared in the same way, and only those features
available in both datasets were used. The results
are presented in Figure 1 and Figure 2. Evaluating
the MIMIC-III-based model with the COPRA dataset
shows that the model is unable to make predictions on
a new independent dataset.
In this study, we compare both datasets to in-
vestigate possible differences in the data that can
explain the failure of generalization. We evaluate
whether there are minor differences between datasets
that could be solved by algorithmic solutions or if
generalization is a challenge that needs to be solved
by more complex solutions.
2 RELATED WORK
There is currently a vast amount of research regarding
machine learning in healthcare, but research into real-
world applications and arising challenges due to the
need for generalization is limited.
(Chekroud et al., 2024) evaluated a prediction
model for schizophrenia patients utilizing multiple
clinical trials and showed good accuracy for intra-trial
evaluations but failed for inter-trial evaluations. They
concluded that findings based on a single dataset pro-
vide limited insight into the general and future perfor-
mance of a model.
(Tonneau et al., 2023) have evaluated the general-
ization of a machine learning model in the domain
of radiomics. Although generalization results have
been improved by algorithmic solutions for one com-
bination of datasets, another combination of datasets
failed generalizability validations.
(Dexter et al., 2020) evaluated machine learning
generalization using free text laboratory data detec-
tion of specific diagnoses. Results show a signif-
icant decrease of model performance for inter-trial
evaluations, with the area under curve for the re-
ceiver operator characteristic as low as 0.48, indicat-
ing a model performance as good as guessing. They
concluded that “studies showing highly performant
machine learning models for public health analyti-
cal tasks cannot be assumed to perform well when
applied to data not sampled by the model’s train
dataset.”
Current research shows that while well-
performing models can be developed in healthcare,
generalization remains a challenge.
3 METHODS
Our goal was to evaluate the challenges of general-
izing machine learning methods in healthcare. To
achieve this, we conducted a comparative analysis be-
tween the COPRA database, used for evaluation in
(Moazemi et al., 2022) and the MIMIC III database.
The COPRA database includes 5,524 patient records,
while the MIMIC database contains 30,284 patient
records. Our analysis focused on two key aspects:
demographic information and time series data and the
differences of these features between the datasets.
3.1 Patient Demographics
We started with demographic data for age, height, and
weight retrieved from both databases. In this com-
parison, we analyze whether there were any signifi-
cant discrepancies in the distribution of demographics
of the patients. We visualized the demographic dis-
tributions through KDE (Kernel Density Estimation)
plots. KDEs are a smoothed, continuous estimate of
the probability density function that describes the data
to compare distribution shapes and central tendencies
more easily between datasets.
To statistically assess any observed disparities, we
employed the following tests:
Welch’s t-test This test evaluates the significance
of the difference between the means of two datasets.
The t-statistic is calculated as (WELCH, 1947):
t =
¯
X
1
−
¯
X
2
r
s
2
1
n
1
+
s
2
2
n
2
(1)
where
¯
X
1
and
¯
X
2
represent the sample means of the
two datasets, s
2
1
and s
2
2
are the sample variances, and
Challenges of Generalizing Machine Learning Models in Healthcare
255

n
1
and n
2
are the sample sizes of the two datasets.
This formula calculates how many standard devia-
tions the difference between the sample means is, pro-
viding a measure of the significance of the mean dif-
ference.
Kolmogorov-Smirnov. This test assesses whether two
samples come from the same distribution. The K-S
statistic is defined as (Massey, 1951):
D = sup
x
|
F
1
(x) − F
2
(x)
|
(2)
where F
1
(x) and F
2
(x) are the empirical cumulative
distribution functions of the two samples, and sup
x
denotes the supremum over all possible values of x.
The two statistical methods mentioned above are
a supplement to assess whether there are differences
between the distributions (apart from visual examina-
tion). The t-test is specifically designed to compare
the means of two groups and the K-S test compares
whether there are differences in the shape of the dis-
tributions, The K-S test does not assume a specific
distribution (like normality) and can be used with or-
dinal data or when the distribution of the data is un-
known.
3.2 Multivariate Time Series
We focused on Temperature, Oxygenation, Heart
Rate, and ambulatory blood pressure (ABP) for the
time series variables. To compute the distance be-
tween time series, evenly spaced time points are nec-
essary. Measurements are irregular across the original
datasets, hospital stays, features, and across time, as
they span from several minutes to hours. All time se-
ries data is resampled to a 15-minute sampling rate to
minimize the loss of information while regularizing
the time series. 15-minute bins are created through-
out the hospital stay, and the mean value of all values
in each bin is used for resampling.
We compared the time series data following two
complementary strategies:
First Strategy. We extracted all time series data for
each variable (e.g., temperature) from the COPRA
and MIMIC database and computed the following de-
scriptors: mean, standard deviation (std), trend, sea-
sonality, and cycle. These descriptors summarized the
central tendencies and temporal patterns within each
time series and are explained in Figure 3. To facili-
tate comparison, we plotted the distributions of these
descriptors for each variable, similar to our approach
to comparing patient demographics. This visualizes
any possible differences in the distribution of the vital
features and temporal patterns between the datasets.
Figure 3: Decomposition of a 48-hour vital sign time se-
ries into its components: cycle, trend, and seasonality. The
cycle refers to the longer-term oscillations within the data
that occur over an extended period, capturing patterns be-
yond daily or short-term fluctuations. The trend represents
the overall direction or progression of the vital sign data
over time, whether increasing, decreasing, or remaining
constant. Seasonality highlights the recurring, predictable
patterns that repeat at regular intervals within 48 hours. To-
gether, these components help describe the structure of the
time series.
Second Strategy. We compared time series data us-
ing Dynamic Time Warping (DTW). DTW is a ro-
bust method for measuring the similarity between
two time series that may vary in speed or timing.
Given two time series, X = (x
1
, x
2
, ..., x
n
) and Y =
(y_1, y_2, ...., y
m
)where n and m represent the lengths
of the two-time series, DTW calculates an optimal
alignment between these sequences by minimizing
the cumulative distance (Müller, 2007).
The DTW distance is calculated by constructing
an n × m cost matrix C(i, j), where each element
C(i, j)represents the distance between points x
i
and
y
j
and is typically calculated as the squared Euclidean
distance:
C(i, j) = (x
i
− y
j
)
2
The goal is to find a warping path W = (w
1
, w
2
, ...w
L
)
where each w
k
= (i
k
, j
k
) maps indices fromX to Y ,
that minimizes the total cumulative cost:
DTW (X,Y ) = min
W
(
L
∑
k=1
C(w
k
)) (3)
This optimal path minimizes the total distance by
allowing for shifts in time (i.e., stretching and com-
pressing of sequences) to better align the series. The
DTW distance provides a measure of similarity be-
tween the time series, with a lower DTW distance in-
dicating greater similarity.
Due to computational limitations, we computed
the DTW distance for a random subset of the data,
randomly sampling 1000 different data points from
each dataset for each vital feature (Temperature, Oxy-
genation, Heart Rate, and ABP). For example, we
measured the distance between 1000 temperature
time series from COPRA and 1000 temperature time
series from MIMIC.
HEALTHINF 2025 - 18th International Conference on Health Informatics
256
We created a DTW distance matrix between all
time series and plotted a heatmap to visualize any dif-
ferences between and in the two groups. This initial
analysis provided a broad and unbiased overview of
the similarities and differences between the datasets.
However, a high number of null values might impact
the DTW comparison. The reason is that all null
values are interpolated before calculating DTW dis-
tances, potentially misleading comparisons by com-
paring interpolation values (in case of many contin-
uous null values, e.g., a pattern line) rather than ac-
tual time series patterns. Recognizing this can lead to
misleading results due to the high proportion of null
values, we conducted a secondary analysis. This anal-
ysis involved filtering out time series with more than
60 percent null values. In addition, we excluded time
series with fewer than 48 data points. We chose this
threshold because shorter time series might not cap-
ture enough temporal variation, something that is es-
sential for a meaningful DTW comparison.
4 RESULTS
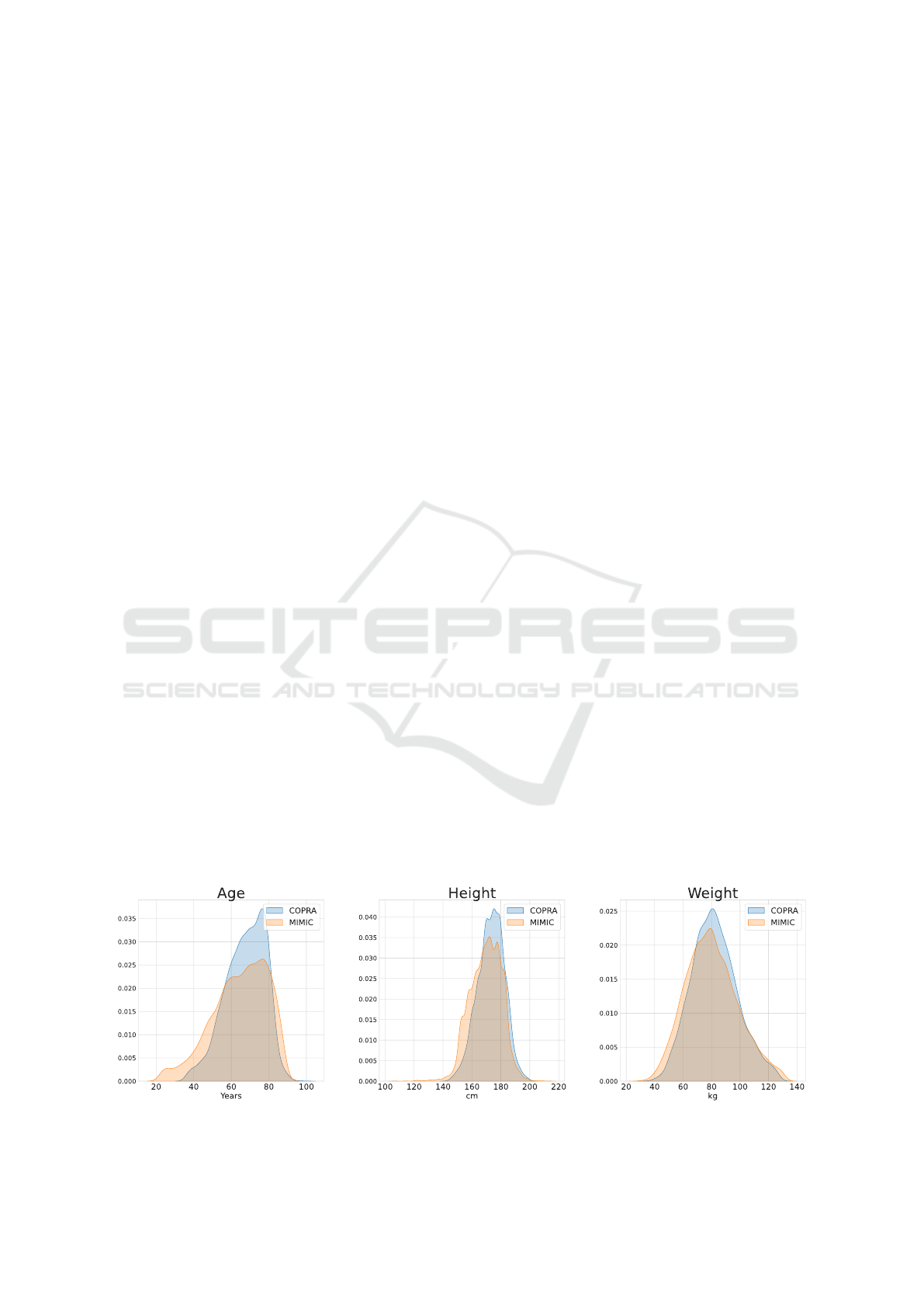
Figure 4 shows the comparison of demographic dis-
tributions of the variables age, height, and weight be-
tween COPRA and MIMIC datasets. The distribu-
tions of both datasets are overlapped to distinguish
the differences better. The distributions are approx-
imately normally distributed, although the age distri-
bution shows a skew towards higher values.
Table 1 shows the results of the t-test and k-test.
It shows significant differences between the COPRA
and MIMIC datasets for age, height, and weight. The
large t-statistics tell us that these differences are not
just by chance; they are substantial. In particular,
height and weight show even bigger differences than
age. It is also important to note that the high t-values
are influenced by the large number of patient records
we compared. With such big groups, even slight dif-
ferences can become statistically significant, so while
these differences are fundamental, the large sample
sizes make them stand out even more.
Figure 5 compares the time series descriptors for
Temperature, Oxygen, Heart Rate, and Arterial Blood
Pressure (ABP) between the COPRA and MIMIC
datasets. For each variable, violin plots display the
distribution of five key descriptors: mean, standard
deviation, trend, seasonality, and cycle. This visual
comparison helps us assess how consistent or variable
the time series data are across the two datasets.
The violin plots show the differences between
the two datasets across various descriptors. One of
the most striking differences is seen in the season-
ality of Temperature, where the median values for
COPRA and MIMIC are entirely different. More-
over, the cycles of the same variable are uniform in
MIMIC, whereas the COPRA dataset displays a pat-
tern with two distinct modes. These temperature vari-
ations could be due to differences in data collection
or processing methods used in the two datasets, or
they might reflect differences in patient populations
or conditions.
Figure 6 shows heatmaps of the distance between
time series computed via DTW. A majority of the
time series consists of mostly missing values due to
the resampling method and missing data. To calcu-
late the DTW, a linear interpolation is used to replace
the missing values. Due to computational limitations,
a subset of 2000 points from each dataset was used.
Brighter colors indicate a higher distance. The tem-
perature heatmap shows clearly that the distances in
the COPRA dataset are the lowest, and the distances
between COPRA and MIMIC are slightly higher than
the distances in the MIMIC dataset. There is no clear
visual difference for the other vital parameters.
To corroborate the visual results, the median dis-
tances are computed. The results are shown in Table
3. For Temperature and Oxygenation, the distance be-
Figure 4: Comparison of demographic distributions.
Challenges of Generalizing Machine Learning Models in Healthcare
257
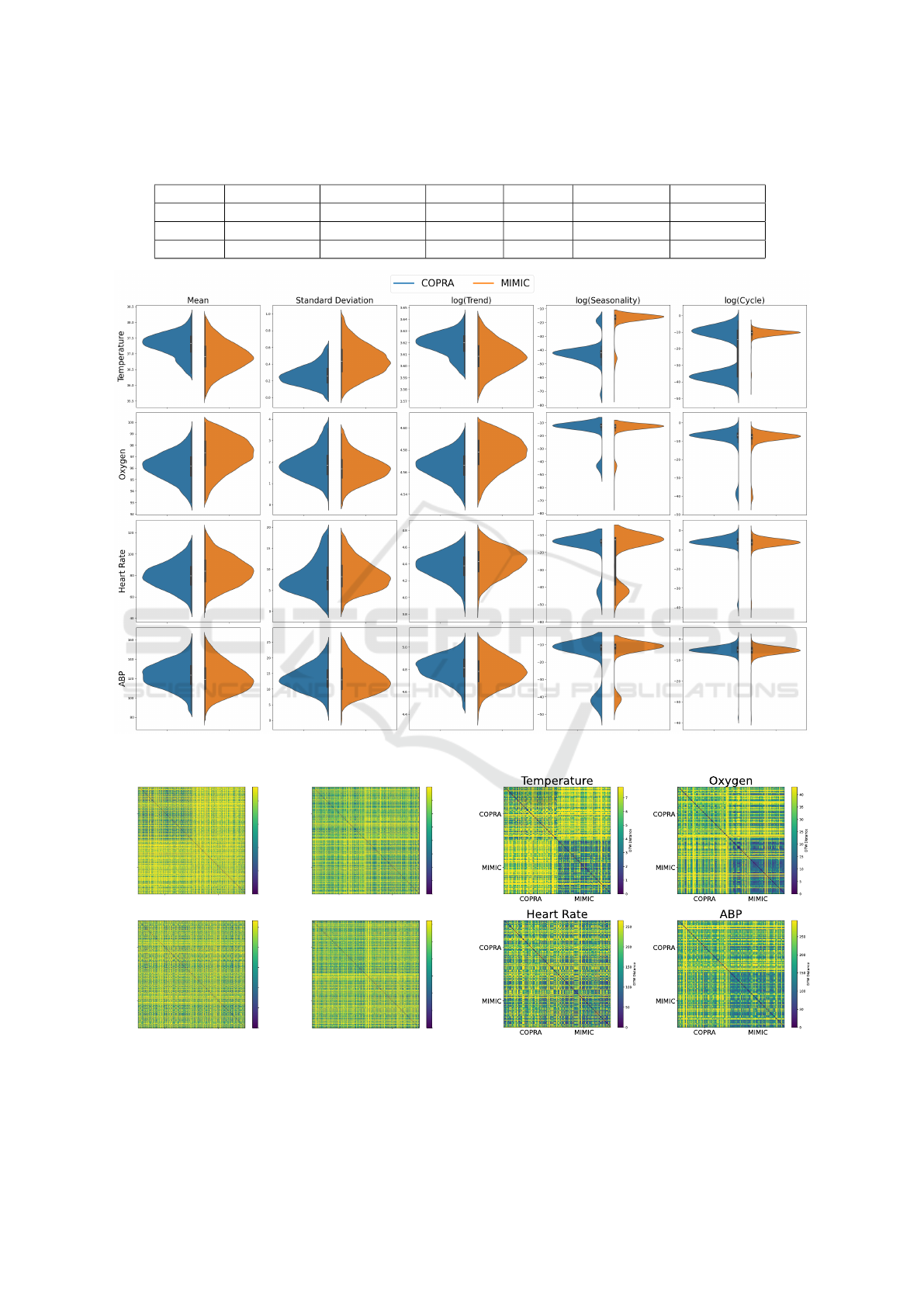
Table 1: Results of t-tests and Kolmogorov-Smirnov test comparing Demographic Variables Between the COPRA and MIMIC
Databases. All tests passed.
Variable Copra Mean MIMIC Mean t-Statistic p-Value K-S Statistic K-S p-Value
Age 66.83 62.02 13.39 < 0.001 0.26 < 0.001
Height 160.00 139.38 26.99 < 0.001 0.28 < 0.001
Weight 79.58 67.66 22.23 < 0.001 0.19 < 0.001
Figure 5: Comparison of time series descriptors across different variables.
COPRA MIMIC
COPRA
MIMIC
Temperature
COPRA MIMIC
COPRA
MIMIC
Oxygen
COPRA MIMIC
COPRA
MIMIC
Heart Rate
COPRA MIMIC
COPRA
MIMIC
ABP
0
2
4
6
8
DTW Distance
0
5
10
15
20
25
30
35
DTW Distance
0
50
100
150
200
250
DTW Distance
0
50
100
150
200
250
300
DTW Distance
Figure 6: The DTW distances for vital values with 2000
data points from each data set.
Figure 7: The DTW distances for vital values. Only data
points with a limited number of missing values are depicted.
HEALTHINF 2025 - 18th International Conference on Health Informatics
258

tween COPRA and MIMIC is the highest, whereas,
for the other two features, the distance in the MIMIC
dataset is the highest. Due to the missing values, some
differences in the data might be unclear, as there can
be no new information obtained by comparing miss-
ing values against missing values.
Table 2: The median distances within and between datasets
with only data points with a limited number of missing val-
ues. The largest median distance for each feature is in bold.
COPRA MIMIC Mixed
Temperature 5.94 3.98 7.42
Oxygenation 32.16 18.15 33.38
Heart Rate 191.90 139.96 179.11
ABP 215.29 143.45 202.15
Table 3: The median distances within and between datasets.
The largest median distance for each feature is in bold.
COPRA MIMIC Mixed
Temperature 6.13 8.82 9.38
Oxygenation 26.63 26.60 29.23
Heart Rate 177.72 190.94 187.14
ABP 207.16 232.45 224.33
To obtain some more visual and precise numerical
results, we performed this analysis with time series
that consist of less than 60% missing values and at
least 12 hours of data. The results are shown in Fig-
ure 7 and Table 2. The higher inter-dataset distance is
more clearly visible for both Oxygenation and Tem-
perature. In Table 3, the effect of removing null values
can clearly be seen; the DTW mean distance within
the MIMIC group decreases, indicating that a large
number of null values can distort the results of DTW
calculations. On the other hand, the distances within
COPRA did not change significantly, suggesting that
the analyzed COPRA dataset has higher-quality data
with fewer null values.
5 DISCUSSION
Differences in data are also evident in the distri-
bution of vital features, which cannot be attributed
solely to patient demographics and may instead stem
from variations in measurement techniques and clin-
ical protocols. This discrepancy is highlighted in our
DTW analysis, where Temperature and Oxygenation
show particularly high distances between the datasets.
(Lin et al., 2019) identify Oxygenation and Temper-
ature as critical for predicting patient readmissions
and suggest that substantial differences in these fea-
tures could impair model generalization. This find-
ing indicates that there is no "free lunch" in machine
learning: models learn from and fit to the data distri-
bution they are trained on, and they cannot generally
make reliable predictions for truly out-of-distribution
data. This also shows in the results of (Moazemi
et al., 2022): Slight differences in data would result
in less predictive performance, which could be im-
proved, e.g., by imputation methods. However, such
a failure to make correct predictions indicates that the
problem needs to be solved a step before: in data col-
lection and the construction of the model architecture.
To build general model architectures, these need
to be built with limitations of future datasets in mind -
if the architecture relies on relatively high availability
and quality of the data (which is available with pub-
lic datasets like MIMIC) it might not work for less
established datasets.
Another challenge arises from differences in miss-
ing values and sampling rates within and across
datasets. These variations complicate dataset com-
parisons and impact model performance. If a criti-
cal feature in a new dataset has a low sampling rate
or a significant amount of missing data, it is unrealis-
tic to expect the model to utilize the data effectively,
even if it captures relationships in a more complete
dataset. Although algorithmic solutions such as impu-
tation can be helpful, they are no substitute for high-
quality data. Ultimately, creating more generalizable
models will require improvements in data quality and
availability.
6 CONCLUSION
By examining MIMIC-III and COPRA datasets, we
found significant differences in demographic distribu-
tion and vital signs, which are likely to impact model
performance, limiting the efficacy of models trained
on one dataset when applied to another. These find-
ings highlight the need for models that not only pre-
dict accurately within a controlled setting but also
adapt to the diverse and evolving nature of real-world
healthcare data. Healthcare data is uniquely challeng-
ing due to its heterogeneity and how commonly data
is missing (Wells et al., 2013). Differences in clinical
practices, patient care protocols, and patient demo-
graphics between hospitals contribute to disparities in
datasets, which can alter model predictions and com-
promise patient outcomes. Such variations between
datasets make it clear that achieving generalizabil-
ity requires more than just refining model architec-
tures. Algorithmic approaches like data imputation
and transfer learning can mitigate these issues but are
not sufficient for data with stark differences. This im-
pacts machine learning in the domain of healthcare
Challenges of Generalizing Machine Learning Models in Healthcare
259

more than other domains, such as general natural lan-
guage processing, where large, well-curated datasets
are available.
As such, developing machine learning applica-
tions for healthcare needs more consideration. De-
veloping a machine learning model that makes cor-
rect predictions for one dataset might not be enough
to build general real-world applications. Overall, to
ensure the development and integration of machine
learning into healthcare applications, more collabo-
ration, more standards, and more data collection are
needed.
ACKNOWLEDGMENT
This research is funded by the German Federal Min-
istry of Education and Research (BMBF) under the
grant numbers 16KISR003 and 13GW0598C and by
the Susanne Bunnenberg Heart Foundation. We are
grateful for the funding received.
REFERENCES
Al-Zaiti, S., Martin-Gill, C., Zègre-Hemsey, J., Bouzid, Z.,
Faramand, Z., Alrawashdeh, M., Gregg, R., Helman,
S., Riek, N., Kraevsky-Phillips, K., Clermont, G., Ak-
cakaya, M., Sereika, S., Dam, P., Smith, S., Birnbaum,
Y., Saba, S., Sejdic, E., and Callaway, C. (2023). Ma-
chine learning for ecg diagnosis and risk stratification
of occlusion myocardial infarction. Nature Medicine,
29:1–10.
Alaa, A. M., Bolton, T., Di Angelantonio, E., Rudd, J. H. F.,
and van der Schaar, M. (2019). Cardiovascular disease
risk prediction using automated machine learning: A
prospective study of 423,604 uk biobank participants.
PLOS ONE, 14(5):1–17.
Chekroud, A. M., Hawrilenko, M., Loho, H., Bondar,
J., Gueorguieva, R., Hasan, A., Kambeitz, J., Cor-
lett, P. R., Koutsouleris, N., Krumholz, H. M., Krys-
tal, J. H., and Paulus, M. (2024). Illusory gener-
alizability of clinical prediction models. Science,
383(6679):164–167.
Dexter, G. P., Grannis, S. J., Dixon, B. E., and Kasthuri-
rathne, S. N. (2020). Generalization of Machine
Learning Approaches to Identify Notifiable Condi-
tions from a Statewide Health Information Exchange.
AMIA Joint Summits on Translational Science pro-
ceedings. AMIA Joint Summits on Translational Sci-
ence, 2020:152–161.
Johnson, A., Pollard, T., and Mark, R. (2023). Mimic-iii
clinical database.
Kumari, J., Kumar, E., and Kumar, D. (2023). A structured
analysis to study the role of machine learning and deep
learning in the healthcare sector with big data analyt-
ics. Archives of Computational Methods in Engineer-
ing, 30(6):3673–3701.
Lin, Y.-W., Zhou, Y., Faghri, F., Shaw, M. J., and Campbell,
R. H. (2019). Analysis and prediction of unplanned
intensive care unit readmission using recurrent neu-
ral networks with long short-term memory. PloS one,
14(7):e0218942.
Massey, F. J. (1951). The kolmogorov-smirnov test for
goodness of fit. Journal of the American Statistical
Association, 46(253):68–78.
Moazemi, S., Kalkhoff, S., Kessler, S., Boztoprak, Z., Het-
tlich, V., Liebrecht, A., Bibo, R., Dewitz, B., Licht-
enberg, A., Aubin, H., et al. (2022). Evaluating a
recurrent neural network model for predicting read-
mission to cardiovascular icus based on clinical time
series data. Engineering Proceedings, 18(1):1.
Müller, M. (2007). Dynamic time warping. Information
retrieval for music and motion, pages 69–84.
Panch, T., Mattie, H., and Celi, L. A. (2019). The “in-
convenient truth” about ai in healthcare. NPJ digital
medicine, 2(1):1–3.
Shapiro, D., Lee, K., Asmussen, J., Bourquard, T., and
Lichtarge, O. (2023). Evolutionary action–machine
learning model identifies candidate genes associated
with early-onset coronary artery disease. Journal of
the American Heart Association, 12(17):e029103.
Tonneau, M., Phan, K., Manem, V. S. K., Low-Kam, C.,
Dutil, F., Kazandjian, S., Vanderweyen, D., Panasci,
J., Malo, J., Coulombe, F., Gagné, A., Elkrief, A.,
Belkaïd, W., Di Jorio, L., Orain, M., Bouchard, N.,
Muanza, T., Rybicki, F. J., Kafi, K., Huntsman, D.,
Joubert, P., Chandelier, F., and Routy, B. (2023). Gen-
eralization optimizing machine learning to improve
CT scan radiomics and assess immune checkpoint
inhibitors’ response in non-small cell lung cancer:
a multicenter cohort study. Frontiers in Oncology,
13:1196414.
WELCH, B. L. (1947). The generalization of ‘student’s’
problem when several different population varlances
are involved. Biometrika, 34(1-2):28–35.
Wells, B. J., Chagin, K. M., Nowacki, A. S., and Kattan,
M. W. (2013). Strategies for Handling Missing Data
in Electronic Health Record Derived Data. eGEMs,
1(3):1035.
HEALTHINF 2025 - 18th International Conference on Health Informatics
260
