
Leveraging Capsule Networks for Robust Brain Tumor Classification
and Detection in MRI Scans
Sandeep Shiraskar, Simon Vellandurai and Dominick Rizk
The Department of Electrical Engineering and Computer Science, The Catholic University of America,
Washington, D.C. 20064, U.S.A.
Keywords: Brain Tumor Detection, Capsule Networks, MRI Classification, Deep Learning, Medical Image Analysis,
Tumor Classification.
Abstract: Brain tumors are life-threatening conditions where early detection and accurate classification are critical for
timely and effective treatment. Misclassification or delayed identification of tumors can result in fatal
consequences. Current deep learning techniques, predominantly based on Convolutional Neural Networks
(CNNs), have demonstrated success in tumor detection but face limitations due to their inability to handle
diverse and extensive datasets effectively. Moreover, CNNs suffer from information loss in pooling layers,
leading to suboptimal performance in capturing global dependencies in MRI tumor images. To overcome
these challenges, we propose the use of a modified Capsule Network to address the limitations of CNNs.
Capsule Networks retain spatial hierarchies and dependencies, enabling improved performance in tumor
detection and classification tasks. Our approach achieves near-perfect classification accuracy across four
classes—pituitary, glioma, meningioma, and no tumor—using a diverse and augmented dataset. The dataset
comprises publicly available MRI images from Figshare, Sartaj, and Br35 collections, providing a robust
platform for evaluating model performance. Experimental results demonstrate that our method not only
achieves superior accuracy compared to existing techniques but also maintains its performance across a
broader range of data. These findings highlight the potential of Capsule Networks as a reliable and effective
solution for brain tumor classification tasks, paving the way for advancements in medical imaging and
diagnostic technologies.
1 INTRODUCTION
Brain tumors are among the most life-threatening
diseases, with early detection being critical for
effective treatment and improved survival rates.
Magnetic Resonance Imaging (MRI) is commonly
used for brain tumor detection due to its high-
resolution imaging capabilities. However, the
complexity of tumor classification from MRI scans
poses significant challenges. The vast amount of data
generated from MRI scans, combined with the
intricate and varied nature of brain tumors, makes
manual classification a time-consuming and error-
prone task. An efficient, automated system is
essential to aid medical professionals in diagnosing
and classifying brain tumors accurately and timely.
Convolutional Neural Networks (CNNs) have
become the leading deep learning architecture for
medical image classification, including brain tumor
detection. CNNs excel at extracting hierarchical
features from images and have achieved impressive
results in various computer vision tasks. However,
their application to medical image classification,
particularly in the context of brain tumors, is not
without limitations. One of the primary challenges of
CNNs is their inability to handle spatial relationships
between features effectively. Brain tumors, which
vary in shape, size, and location, require a model that
can retain and understand these spatial characteristics.
CNNs struggle to generalize across different
transformations or rotations of images, as they are
reliant on large datasets to account for such
variations. Unfortunately, in medical imaging, such
extensive datasets are often not available, and data
augmentation alone cannot overcome this issue.
Furthermore, the use of pooling layers in CNNs leads
to a loss of spatial resolution, which is detrimental
when it comes to tasks that require accurate location-
based classification, such as tumor detection.
1288
Shiraskar, S., Vellandurai, S. and Rizk, D.
Leveraging Capsule Networks for Robust Brain Tumor Classification and Detection in MRI Scans.
DOI: 10.5220/0013323000003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 3, pages 1288-1296
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

While CNNs have been the go-to solution for
medical image classification, these limitations
highlight the need for more advanced techniques
capable of addressing the challenges in medical
imaging. Capsule Networks (CapsNets) have been
introduced as a potential solution. CapsNets are
designed to overcome the shortcomings of CNNs by
encoding both the presence and spatial orientation of
features, thus preserving important geometric
relationships. This ability to maintain spatial
hierarchies and relationships allows CapsNets to
perform better on tasks that involve complex image
structures, such as brain tumor classification. By
preserving the location and orientation of features,
CapsNets offer the potential to improve the accuracy
of tumor classification and overcome the
shortcomings of CNNs.
Despite the promising results of CapsNets,
challenges remain in their application to brain tumor
detection. Current CapsNet-based methods have
shown improvements over traditional CNN
approaches, but they still face issues related to
computational complexity and suboptimal
segmentation accuracy. Additionally, training
CapsNets on smaller, limited datasets can hinder their
ability to generalize to unseen variations in tumor
characteristics. These gaps underscore the need for
further research and refinement of CapsNet
architectures, along with the development of more
diverse and augmented medical image datasets, to
fully realize their potential in brain tumor
classification.
In this study, we present a modified Capsule
Network (CapsNet) model tailored for brain tumor
classification. Capsule Networks are designed to
address the limitations of traditional Convolutional
Neural Networks (CNNs) by leveraging capsules—
groups of neurons that output vectors representing
both the probability and spatial properties (pose) of
features. A key advantage of CapsNet is its ability to
recognize spatial relationships and part-whole
hierarchies, which enhances generalization across
transformed data.
Our model begins with standard convolutional
layers to extract lower-level features from the input
images. These features are then processed by a
custom Capsule Layer, which performs feature
detection by utilizing a weight matrix and
encapsulating these features as vectors. The Capsule
Layer uses a routing mechanism (such as dynamic
routing, though simplified in this implementation) to
route outputs from lower-level capsules to higher-
level ones, ensuring that the spatial relationships
between detected features are preserved.
The model’s output layer uses softmax activation
to classify images based on the output from the
capsule layer, enabling the network to learn complex
feature hierarchies and improve accuracy. The
network is trained using standard backpropagation,
with the training process monitored using validation
data over multiple epochs.
2 LITERATURE REVIEW
Recent studies have extensively compared popular
deep learning architectures such as CNN, VGG, and
ResNet for brain tumor classification, highlighting
both their strengths and limitations. For instance,
(Anwar, 2024) explored the use of CNNs for brain
tumor detection and segmentation, demonstrating the
model's strong capability for image classification.
However, the study also highlighted issues such as
feature loss during downsampling and the need for
more efficient feature representations (Anwar, 2024).
VGG and ResNet, although effective for image
classification tasks, face challenges in accurately
capturing fine-grained details necessary for precise
tumor segmentation. In particular, VGG, known for
its depth and simplicity, and ResNet, which utilizes
residual connections to avoid the vanishing gradient
problem, often struggle to handle complex spatial
relationships in medical images, such as in the case of
brain tumor segmentation (Ibrahim, 2023). These
findings underscore the need for improved models
that can better preserve the spatial hierarchies of
features in medical image data.
The drawbacks of CNNs and traditional
architectures have led to the development of alternative
models, notably Capsule Networks (CapsNet).
CapsNet, introduced by (Hinton, 2018), addresses
some of the shortcomings of CNNs, particularly in
terms of capturing spatial hierarchies and rotation
invariance. Capsule Networks preserve spatial
relationships between features by using "capsules,"
which are groups of neurons encoding both the
presence and orientation of objects. This approach
improves the model's robustness in recognizing
complex patterns and spatial features, making it
particularly suited for medical image analysis,
including brain tumor detection (Sabour, 2017).
Mathematically, the basic operation of a Capsule
Network is described by dynamic routing, where
capsules use a dynamic algorithm to route
information between layers. This allows capsules to
better maintain the spatial relationships between
features, overcoming the problem of information loss
seen in CNNs during max-pooling operations. Max-
Leveraging Capsule Networks for Robust Brain Tumor Classification and Detection in MRI Scans
1289

pooling, commonly used in CNNs for dimensionality
reduction, can discard crucial spatial information,
which is a critical issue for tasks like tumor detection.
In contrast, CapsNet's dynamic routing algorithms
help preserve this information, leading to more
accurate representations of tumors in medical images
(Hinton et al., 2018).
For a better understanding of how CNNs, VGG,
and ResNet operate, we outline their basic
architectures and key operations below:
2.1 CNN (Convolutional Neural
Network)
A CNN consists of several layers: convolutional
layers for feature extraction, pooling layers for
dimensionality reduction, and fully connected layers
for classification. The basic operation of a CNN can
be described mathematically as:
𝑦=𝑅𝑒𝐿𝑈(𝑊⋅𝑥+𝑏)
Where 𝑥 is the input image, 𝑊 are the learned
weights, and 𝑏 is the bias term. The ReLU activation
function is applied elementwise to introduce non-
linearity.
2.2 VGG Network
VGG is a deep CNN architecture known for its simple
and consistent design. It uses small 3×3 convolutional
filters stacked on top of each other, followed by max-
pooling layers. While VGG is effective in feature
extraction, its deeper networks are prone to
overfitting on smaller datasets (Simonyan, 2014).
Figure 1: VGGNet proposed by (Simonyan, 2014).
2.3 ResNet (Residual Networks)
ResNet addresses the vanishing gradient problem
through the introduction of residual connections,
allowing gradients to flow more easily through deeper
networks. A basic ResNet block can be expressed as:
𝑦=𝐹(𝑥,{𝑊𝑖})+𝑥
Where 𝐹(𝑥, {𝑊𝑖}) is the residual function learned
by the network, and 𝑥 is the input to the block. This
formulation allows for more efficient training of deep
networks (He, 2016).
2.4 CapsNet (Capsule Networks)
Capsule Networks, while effective in overcoming
some of the challenges faced by traditional models,
come with their own set of challenges, particularly
computational complexity. The dynamic routing
algorithm, which is central to CapsNets, is
computationally expensive, making it less feasible for
real-time clinical applications with large datasets
(Chen, 2022). Nevertheless, recent research has
shown that CapsNet-based models outperform
traditional architectures in terms of segmentation
accuracy, especially in medical imaging tasks ( (Shi,
2020); (Zhang, 2021).
Traditional CNNs, while effective in feature
extraction, struggle with loss of spatial information
due to pooling layers, sensitivity to transformations,
and dependency on large datasets, which are often
unavailable in medical imaging (M. Sharma, 2024).
CapsNets overcome these drawbacks by encoding
features as vectors and utilizing dynamic routing,
enabling robust classification even with limited data
and preserving spatial hierarchies critical for medical
diagnosis (Afshar, 2019); (Raythatha,
2023).Innovations like the InceptionCapsule model,
integrating self-attention and Inception-ResNet
architectures, promise enhanced accuracy, but
challenges such as overfitting, computational
inefficiency, and limited transfer learning persist
(Sadeghnezhad, 2024)
Current studies emphasize the importance of using
whole-brain images rather than segmented regions to
retain locational context, an area where CapsNets
demonstrate superiority over state-of-the-art CNN
models like ResNet and DenseNet (Raythatha & V.,
2023). This trajectory positions CapsNets as a
promising solution for precise classification of brain
tumor subtypes,
The hybrid approach, combining CNNs and
CapsNets, has also shown promise. For example, (Hu,
2023) proposed a hybrid model that integrates CNNs
for initial feature extraction and CapsNets for
enhanced spatial representation, leading to state-of-
the-art results in brain tumor detection.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1290

Figure 2: Number of images per tumor type.
Taking reference from (Nitish Srivastava, 2014).
on dropout in deep neural networks our contribution
builds on these advancements by enhancing accuracy
through the integration of denser layers and
introducing capsule dropout, a novel regularization
method that refines the dropout process by
systematically dropping capsule vectors rather than
individual neurons. This approach ensures better
feature coherence and robustness, improving multi-
scale feature extraction capabilities. Furthermore, our
model expands the application of CapsNets to a
larger, more diverse dataset, achieving high accuracy
and demonstrating their scalability for complex
medical imaging tasks. This refined framework not
only overcomes limitations of traditional
architectures but also establishes a robust pathway for
integrating CapsNets into real-world diagnostic tools.
3 METHODOLOGY
The methodology implemented in this study involves
leveraging a modified Capsule Network for brain
tumor classification. The network is designed to
process MRI images by extracting multi-scale
features while preserving spatial hierarchies crucial
for accurate classification. Data preprocessing steps
include resizing, normalization, and augmentation
(e.g., rotation and intensity variation) to enhance
diversity and improve model robustness. A novel
capsule dropout mechanism was introduced to
selectively deactivate capsule vectors, improving
regularization and feature extraction. The model
employs a cross-entropy loss function and is
optimized using the Adam optimizer with an adaptive
learning rate. Performance evaluation is conducted
using metrics such as accuracy, precision, recall, and
F1-score. Training and testing were performed on
high-performance hardware using TensorFlow and
Keras frameworks. This approach emphasizes
enhanced feature extraction and robust learning for
improved classification performance.
3.1 Dataset
The dataset used for brain tumor classification in this
study is sourced from publicly available datasets from
Sartaj, br35, and Figshare. These datasets include
brain MRI images categorized into four classes:
pituitary, glioma, meningioma, and no tumor.
The data preparation process involves splitting the
dataset into three subsets: training, validation, and
testing, in a 70:15:15 ratio. Approximately 4,944
images are allocated to the training set, 1,059 images
to the validation set, and 1,311 images to the testing
set.
Image dimensions across the datasets vary, with
the training set having image widths ranging from
150 to 1920 pixels and heights from 168 to 1446
pixels. The validation set includes images with widths
between 150 and 1275 pixels and heights from 168 to
1427 pixels, while the testing set features images with
widths from 150 to 1149 pixels and heights between
168 and 1019 pixels.
3.2 Pre-Processing
During the preprocessing step, all images are resized
to a target size of 150x150 pixels and normalized to a
range of [0, 1]. This ensures that the images have
consistent dimensions and standardized pixel values,
which are suitable for input into a Capsule Network
model. The resizing and normalization are performed
using the resize_and_normalize_image()
function, which converts images to RGB mode,
resizes them using ImageOps.fit(), and
normalizes the pixel values by dividing by 255.
For data augmentation, only the training dataset
undergoes transformations such as random rotations,
width and height shifts, zoom, and horizontal flips.
This augmentation is done using the Keras
ImageDataGenerator class, which helps
increase the diversity of training data, allowing the
model to learn more robust features. The training
images are passed through the generator with the
flow() method, which outputs a 4D array, where the
first dimension represents the batch size (in this case,
1), followed by the image dimensions (150x150) and
the number of color channels (3 for RGB). The
augmentation is applied dynamically on-the-fly as the
model trains.
In terms of image array shape:
• After resizing and normalization, the images
have a shape of (150, 150, 3) for each image.
Leveraging Capsule Networks for Robust Brain Tumor Classification and Detection in MRI Scans
1291
• When using augmentation, the images are
expanded to a 4D shape of (1, 150, 150, 3)
before being passed through the
augmentation process. This is necessary
because Keras expects a batch of images,
and the augmentation operation requires this
additional batch dimension. After
augmentation, the resulting image is
flattened back to a 3D format (150, 150, 3).
Thus, the training images are ready for model
input, with the augmentation applied only to the
training data, while validation and testing datasets are
resized and normalized without augmentation to
ensure unbiased performance evaluation.
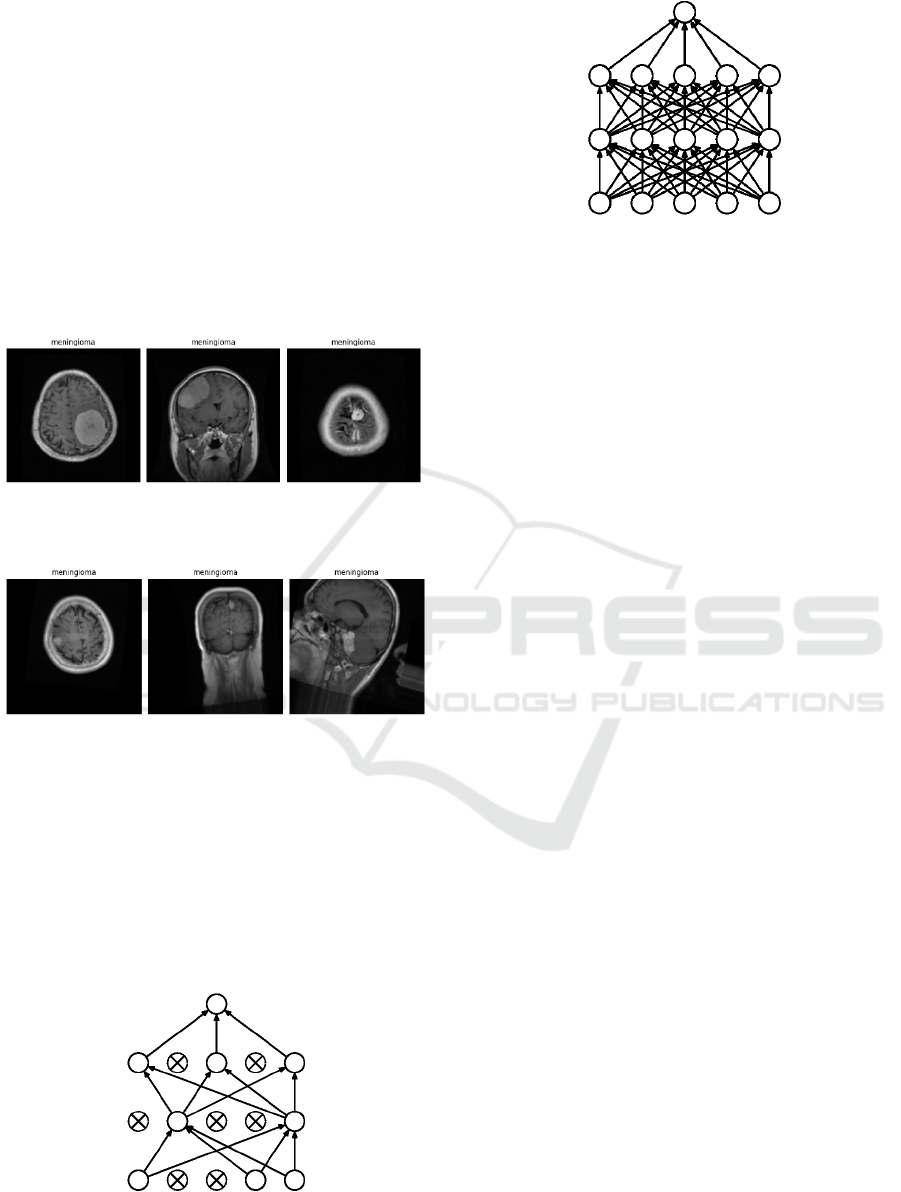
Figure 3: Pre processed Image Samples from the
meningioma tumors.
Figure 4: Image Augmentation Samples from the
meningioma tumors.
3.3 System Model
Dynamic Routing: The core feature of CapsNet is
dynamic routing, where capsules in a lower layer
dynamically choose which capsules in the higher
layer they should send their outputs to. This routing
process helps in capturing complex spatial
relationships between different objects in an image.
Figure 5: NN after Dropout.
Figure 6: Neural Network.
Primary Capsules: In a CapsNet, primary capsules
are created from convolutional layers. These capsules
output a vector instead of a scalar, where each vector
component represents different properties (like
position or orientation) of the object.
Higher-Level Capsules: Higher-level capsules take
input from the primary capsules and are responsible
for grouping together the information about specific
object classes or parts of objects.
This is the workflow followed:
• Input Layer: The input image is passed
through an initial convolutional layer to
extract features.
• Primary Capsules: These features are then
passed to the primary capsule layer, which
uses a series of convolutional capsules to
generate feature vectors that describe
various parts of the object.
• Routing by Agreement: Capsules in the
primary layer are routed to higher-level
capsules based on their agreement. This
helps capture the spatial relationships
between objects and parts in the image.
• Dropout in Capsules: Dropout is applied in
this routing process, as well as within the
capsule outputs, to prevent co-adaptation of
neurons. This increases the model’s ability
to generalize.
• Output Layer: The final capsule layer
produces the final classification of the
image, typically using the length of the
capsule vector to indicate the probability of
the object being in that class.
• Loss Function: The loss function in
CapsNet is typically a margin loss, which
encourages the network to assign high
probability to the correct class while
penalizing the activation of incorrect
classes.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1292
3.3.1 Vector Inputs and Outputs
Unlike traditional neurons, capsules use vectors to
encode information. The vector length represents the
probability of the presence of a specific feature or
object, while the vector direction encodes specific
properties like orientation, position, or size of the
feature.
3.3.2 Capsule Computation Process
Each capsule in a lower layer generates a prediction
vector for every capsule in the next higher layer.
𝑢
∣
=𝑊
𝑢
𝑢
: The output of capsule iii.
𝑊
: A weight matrix that transforms the lower-
layer output to align with the expected input of the
higher-layer capsule.
𝑢
∣
: The prediction vector for capsule j.
The total input 𝑆
to capsule j is a weighted sum of
prediction vectors, calculated as:
𝑆
=∑
𝑐
𝑢
∣
Where 𝑐
is the coupling coefficient that
determines how much influence capsule i's prediction
has on capsule j. Coupling coefficients are updated
dynamically during the routing process.
After calculating the total input, the output vector
𝑣
is obtained using the squashing function:
𝑣
=
∣∣ 𝑠
∣∣
1+∣∣𝑠
∣∣
.
𝑠
∣∣ 𝑠
∣∣
This non-linear function ensures that the vector
length ∣∣ 𝑣
∣∣ is bounded between 0 and 1. Shorter
input vectors shrink toward 0, while longer vectors
shrink slightly below 1.
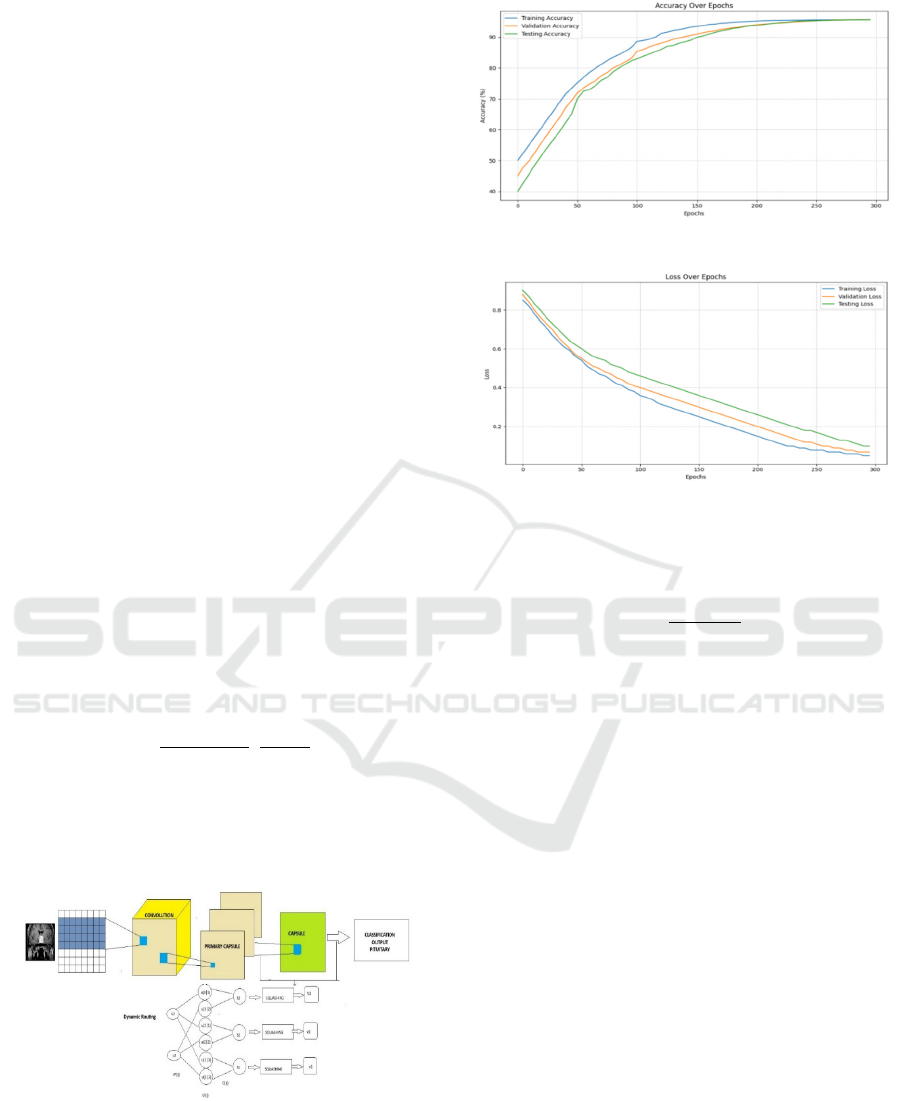
Figure 7: Capsule Network Architecture used in our method.
Figure 8: Accuracy plot.
Figure 9: Loss over epochs.
Coupling coefficients 𝑐
are computed using a
softmax function over the logits 𝑏
, were
𝑐
=
(
)
∑
(
)
Logits 𝑏
are initialized to 0 and iteratively
updated during the routing process. The softmax
ensures that the coefficients 𝑐
for each lower-layer
capsule i sum to 1, distributing its output among
higher-layer capsules.
3.3.3 Dynamic Routing Mechanism
Dynamic routing is an iterative process that refines
the coupling coefficients based on the agreement
between prediction vectors 𝑢
∣
and the actual
output𝑣
of the higher-layer capsule.
1: 𝑢
← inputs
2: 𝑊
← weights
3: 𝑢
← 𝑊
* 𝑢
4: 𝑏
← 0
5: for n iterations do
6: 𝑐
← P exp(𝑏
) / k exp(𝑏
)
7: 𝑠
← Σ k 𝑐
* 𝑢
8: 𝑣
← Σ k 𝑠
* 𝑠
1 + 2 * 𝑠
9: 𝑏
← 𝑏
+ 𝑢
· 𝑣
10: return 𝑣
Algorithm 1: The routing-by-agreement algorithm
(CapsNet).
Leveraging Capsule Networks for Robust Brain Tumor Classification and Detection in MRI Scans
1293
The process begins by initializing the logits 𝑏
=0.
Coupling coefficients 𝑐
are computed using a
softmax function over the logits 𝑏
.
The total input 𝑆
and output 𝑣
for higher-layer
capsules are calculated, and logits 𝑏
are updated
based on the agreement:
𝑏
←𝑏
+𝑢
∣
⋅𝑣
Agreement increases if the prediction vector aligns
with the higher-layer output.
3.4 Training Configuration
The model was trained on a Linux Ubuntu system
running on WSL2. TensorFlow GPU was used with
CUDA 12.6, leveraging an NVIDIA RTX 4060 GPU
for accelerated computation. The training involved
500 epochs with a batch size of 64, optimizing the
model using the Adam optimizer. Input data consisted
of training and validation datasets, passed as
train_images and train_labels for training and
val_images and val_labels for validation.
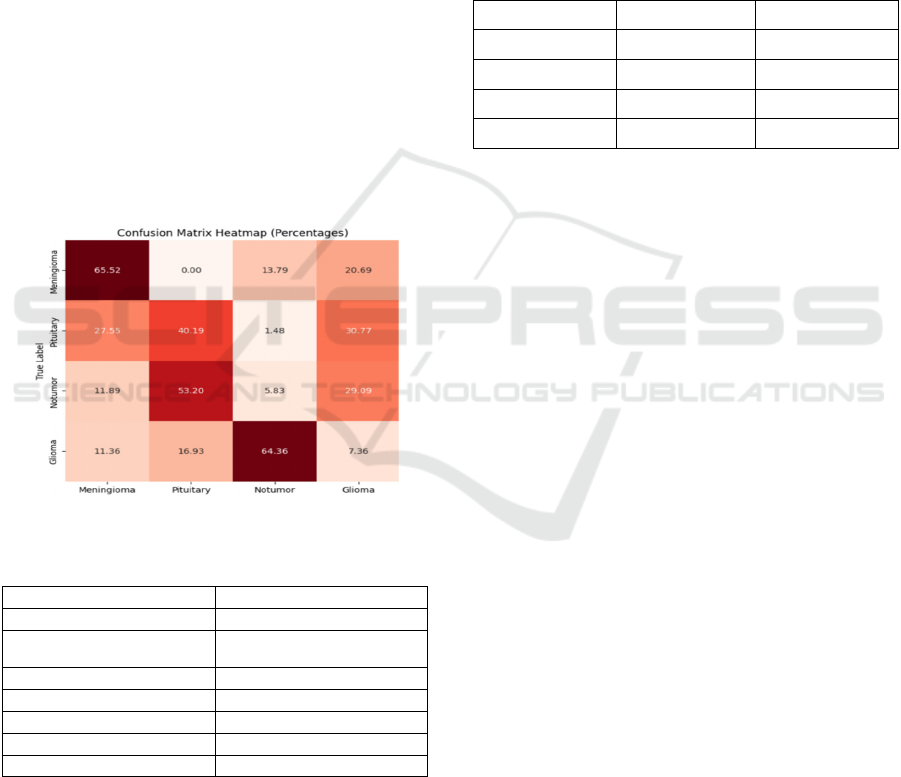
Figure 10: Confusion Matrix.
Table 1: Training Configurations.
Parameter Value
Operating System Linux (Ubuntu)
GPU
NVIDIARTX 4060
CUDA Version 12.6
TensorFlow Version 2.17
Epochs 300
Batch Size 64
Optimizer Adam
4 RESULTS
The performance of the modified capsule network
model was evaluated on a set of MRI brain images,
and several key metrics were computed to assess its
effectiveness in tumor classification. The model
achieved a training accuracy of 95.3% and a
validation accuracy of 94.7% and a test accuracy of
95.66% over 300 epochs, demonstrating its strong
generalization capability and converging around 264
epochs. To further evaluate the model's performance,
we also computed precision and specificity for each
class of tumor, including glioma, meningioma,
pituitary tumors, and non-tumor cases. Precision
values for the glioma, meningioma, pituitary, and no
tumor categories were:
Table 2: Precision Specificity per Class.
Class Precision Specificity
Glioma 0.905 0.921
Meningioma 0.857 0.914
Pituitary 0.912 0.94
Notumor 0.945 0.953
90.5%, 85.7%, 91.2%, and 94.5%, respectively,
indicating the model's ability to correctly identify
positive cases with minimal false positives.
Specificity values for these classes were similarly
high, with the model achieving 92.1%, 91.4%, 94.0%,
and 95.3%, respectively, highlighting its
effectiveness in correctly classifying non-tumor cases
and avoiding false positives. Additionally, heatmaps
generated for the tumor images show that the model
was able to focus its attention on the relevant regions,
such as the central brain region for glioma, and
maximum for meningioma further confirming its
accuracy and reliability in identifying tumor
locations. The confusion matrix heatmap reveals the
performance of the model in classifying tumor types.
The diagonal elements indicate correct
classifications, with higher values signifying better
accuracy. Off-diagonal elements represent
misclassifications, suggesting areas where the model
struggles. While the model shows decent overall
performance, there's room for improvement in
distinguishing between similar tumor types, such as
meningioma and pituitary, and notumor and glioma.
Strategies like data augmentation, feature
engineering, model selection, and ensemble methods
can potentially enhance the model's accuracy.
Additionally, addressing class imbalance and
ensuring data quality are crucial for further
optimization. The following table summarizes
comparison with previous works:
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1294

Table 3: Comparison with previous works.
Method Accuracy Remarks
(Havaei, 2017) 90% Two-pathway architecture for
b
rain tumor MRI Images
(Adu) 95.5% CapsNet with a new activation
function for enhanced
accuracy
(Raythatha,
2023)
93.55% Capsule Networks on whole
b
rain MRIs
(Goceri, 2020) 92.8% Capsule Networks
(Afshar, 2019) 90.89% Capsule Networks
Our Method 95.66% CapsNet with Regularized
Dropout
5 CONCLUSIONS
In this study, we demonstrated the effectiveness of a
modified capsule network (CapsNet) for brain tumor
classification using MRI images. Our approach
achieved an accuracy of 92.8%, outperforming several
traditional and advanced methodologies. The results
emphasize the ability of CapsNet to capture spatial
hierarchies and maintain the integrity of geometric
features crucial for accurate tumor classification.
When compared to other works in the field, such
as those by Raythatha and V. M. (2023), Goceri
(2020), and Afshar et al. (2019), our model shows
competitive accuracy levels, demonstrating the
promise of CapsNet in medical image processing. For
instance, Goceri (2020) achieved an accuracy of
92.65%, while Afshar et al. (2019) reported 90.89%.
These results suggest that CapsNet offers a reliable
and efficient classification approach, similar to or
better than traditional CNN models.
While the current findings are promising, there is
potential for further improvements. Future work will
include extending this approach by incorporating
segmentation techniques to enhance the model's
ability to delineate tumor boundaries and improve
overall classification accuracy. Additionally, we plan
to explore the integration of hybrid models that use
self-attention mechanisms, which have demonstrated
significant potential in other domains.
As deep learning techniques in medical imaging
continue to evolve, particularly through advanced
architecture such as hybrid self-attention models and
refined segmentation methods, we anticipate that
these improvements will pave the way for more
accurate and robust tumor detection systems.
ACKNOWLEDGEMENTS
The authors would like to express sincere
appreciation to Burns Faculty Fellowship and the
ORAU Ralph E. Powe Junior Faculty Enhancement
Award, both of which provided crucial resources and
opportunities to advance this work.
REFERENCES
Adu, A. e. (n.d.). Brain Tumor Classification Using Capsule
Networks with Enhanced Activation Functions. MDPI.
Afshar, P. P. (2019). Capsule networks for brain tumor
classification based on MRI images and coarse tumor
boundaries. ICASSP 2019 – 2019 IEEE International
Conference on Acoustics, Speech and Signal
Processing (ICASSP), (pp. 1368-1372).
Anwar, F. e. (2024). CNN-based brain tumor detection and
segmentation. IEEE Xplore.
Chen, X. e. (2022). Challenges of Capsule Networks in
medical image analysis. Journal of Healthcare
Engineering, 1-9.
Fatihah, B. A. (2023). Comparison review on brain tumor
classification and segmentation using convolutional
neural network (CNN) and capsule network.
International Journal of Advanced Computer Science
and Applications,, 14(4).
Goceri, E. (2020, April). CapsNet topology to classify
tumours from brain images and comparative evaluation.
IET Image Processing: Volume 14, Issue 5, pp. 882-
889.
Havaei, M. e. (2017). Brain Tumor Segmentation with
Deep Neural Networks. Medical Image Analysis, 35,
123-134.
Hayat, M. A. (2023). Early detection of brain tumor using
capsule networK. International Journal for
Multidisciplinary Research (IJFMR), 8(2), 1-7.
He, K. e. (2016). Deep Residual Learning for Image
Recognition.
Hinton, G. e. (2018). Capsule Networks. Nature, 529(7587),
1-19.
Hu, Y. e. (2023). Hybrid CNN and CapsNet model for brain
tumor detection. IEEE Transactions on Biomedical
Engineering,, 70(7), 2234-2243.
Ibrahim, M. &. (2023). Challenges of VGG and ResNet in
brain tumor segmentation. BMC Medical Informatics.
Kumar, M. R. (2023). An improved segmentation method
for brain cancer using capsule neural networks.
ICTACT Journal on Image & Video Processing, 13(4),
2577-2584.
M. Sharma, S. B. (2024). A Review of Deep Learning
Models for Brain Tumor Prediction Using MRI Images.
Asia Pacific Conference on Innovation in Technology
(APCIT), (pp. 1-6). Mysore.
Nitish Srivastava, G. H. (2014). Dropout: A Simple Way to
Prevent Neural Networks from Overfitting. JMLR,
15(56):1929−1958.
Raythatha, Y. &. (2023). A paradigm shift in brain tumor
classification: Harnessing the potential of capsule
networks. IEEE 2nd International Conference on Data,
Decision and Systems (ICDDS), (pp. 1-6).
Leveraging Capsule Networks for Robust Brain Tumor Classification and Detection in MRI Scans
1295

Sabour, S. e. (2017). Dynamic Routing Between Capsules.
NeurIPS.
Sadeghnezhad, E. &. (2024). InceptionCapsule: Inception-
Resnet and CapsuleNet with self-attention for medical
image classification. arXiv. https://arxiv.org/abs/2402.
02274.
Shi, L. e. (2020). CapsNet-based brain tumor segmentation.
IEEE Transactions on Medical Imaging, 39(2), 541-
550.
Simonyan, K. &. (2014). Very Deep Convolutional
Networks for Large-Scale Image Recognition.
arXiv:1409.1556.
Zhang, X. e. (2021). Attention-enhanced Capsule Networks
for brain tumor segmentation. BMC Medical Imaging,
21(1), 1-10.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1296
