
Identifying an Autoinflammatory Syndrome Cohort Using Natural
Language Processing with Electronic Medical Record Data
Maranda Russell
1a
, Aleksander Lenert
2b
, Katherine Liao
3c
, Tianrun Cai
3d
and Sujin Kim
4,* e
1
College of Business, Northern Kentucky University, 1 Louie B Nunn Dr BC206, Highland Heights, KY 41099, U.S.A.
2
Department of Internal Medicine, University of Iowa, 200 Hawkins Dr, Iowa City, IA, U.S.A.
3
Department of Biomedical Informatics, Harvard University, 60 Fenwood Road, Boston, MA, U.S.A.
4
Division of Biomedical Informatics, University of Kentucky, 725 Rose Street, Lexington, KY, U.S.A.
Keywords: Autoinflammatory Syndromes (AIS), Clinical Natural Language Processing (cNLP), Machine Learning
Algorithms, Electronic Medical Records (EMR).
Abstract: Autoinflammatory syndromes (AIS) are rare inflammatory disorders with diverse and severe manifestations,
making their clinical outcomes and phenotypes poorly understood. This study developed and validated
machine learning algorithms incorporating clinical natural language processing (cNLP) and electronic
medical record (EMR) data to identify AIS cases. Patients were filtered using relevant billing codes,
medications, and ICD-9/-10 codes for conditions such as adult-onset Still’s disease, Behcet's disease, and
familial Mediterranean fever. Machine learning models—adaptive lasso penalized logistic regression
(ALASSO), support vector machine (SVM), and random forest (RF)—utilized structured codes and cNLP-
extracted features. Of 206 patients screened, 61 (29.6%) were confirmed AIS cases after manual review. SVM
(AUC=0.954) and RF (AUC=0.948) outperformed ALASSO (AUC=0.94). A total of 44 features, including
ICD codes for arthritis and Behcet's disease and cNLP-derived concepts such as periodic fever, oral lesions,
and colchicine treatment, were predictive of AIS. This study demonstrates the feasibility of combining
structured and unstructured EMR data for AIS identification, providing a scalable framework for phenotyping
rare diseases and advancing outcomes research.
1 INTRODUCTION
Autoinflammatory syndromes (AIS) are rare disorders
defined by an exaggerated inflammatory response,
where local factors at disease-predisposed sites
activate innate immune cells, including macrophages
and neutrophils, leading to target tissue damage
(McGonagle, 2006). Clinically, AIS is characterized
by recurrent episodes of arthritis, rash, fever, and
additional systemic manifestations, significantly
impacting quality of life and leading to disability. AIS
pathogenesis involves the inflammasome and the pro-
inflammatory interleukin-1 (IL-1) and interleukin-18
axes, resulting in rheumatic manifestations
(McGonagle, 2006). Additionally, AIS may lead to
a
https://orcid.org/0000-0001-6405-4807
b
https://orcid.org/0000-0003-2129-3263
c
https://orcid.org/0000-0002-4797-3200
d
https://orcid.org/0000-0002-5893-0169
e
https://orcid.org/0000-0002-7878-4322
*
Corresponding author: sujinkim@uky.edu
comorbidities, such as cardiovascular disease, due to
its shared pathogenic mechanisms with atherosclerosis
(Hintenberger, 2018; Ridker, 2016). If untreated, AIS
can progress to severe complications, including
secondary amyloidosis. However, due to the rarity of
AIS and a lack of well-identified longitudinal cohorts,
the full scope of its clinical outcomes remains poorly
understood. The heterogeneity of AIS presentations
and their episodic nature further complicate timely
diagnosis and management. Advances in
computational approaches, including clinical natural
language processing (cNLP) and machine learning
(ML), offer promising avenues for improving the
identification and study of these rare disorders using
electronic medical record (EMR) data.
Russell, M., Lenert, A., Liao, K., Cai, T. and Kim, S.
Identifying an Autoinflammatory Syndrome Cohort Using Natural Language Processing with Electronic Medical Record Data.
DOI: 10.5220/0013323500003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 867-873
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
867

2 BACKGROUNDS
Building a prospective AIS cohort is challenging and
costly, requiring extensive multicentre collaboration
among expert clinicians, researchers, and patient
advocacy groups. In the short term, leveraging large
datasets from EMRs and administrative healthcare
databases offers a promising approach for AIS cohort
identification, facilitating clinical outcomes research
and translational studies in rheumatic diseases (Hak,
2009; Desai, 2005). One major challenge in AIS
cohort building is accurately identifying and
capturing all AIS cases for epidemiologic and
translational research. While ICD-9 (International
Classification of Diseases, 9th Revision) codes have
traditionally been used to identify rheumatic disease
phenotypes, including rheumatoid arthritis (RA) and
systemic lupus erythematosus, validated algorithms
for accurate AIS identification are currently lacking
(Liao, 2015; Barbhaiya, 2017; Feldman, 2013;
Feldman, 2015, Kim, 2017).
The availability of longitudinal EMRs for
clinical research has proven valuable for
phenotyping rare rheumatic diseases and associated
outcomes (Kim, 2011; Brownstein, 2010; Liao,
2014). Recently, robust algorithms that integrate
structured and unstructured EMR data have
improved phenotyping for conditions such as RA,
outperforming purely coding-based approaches
(Ramirez, 2014; Liao, 2010). These algorithms often
employ cNLP to extract rich clinical data from
narrative notes. cNLP is a computational method
that identifies concepts in clinical text using
linguistic rules, making it particularly useful for
rheumatic diseases like AIS, which have poorly
defined ICD-9/-10 codes and low prevalence (Desai,
2017). Through cNLP, unstructured narrative data
can be transformed into analysable datasets.
Working closely with advanced cNLP and machine
learning algorithms, this study aimed to develop and
validate a preliminary algorithm optimized to
maximize both positive and negative predictive
values for AIS case identification from EMR data.
3 METHODS
3.1 Study Design and Data Collection
This study utilized a modified surrogate-assisted
feature extraction (SAFE) procedure as described by
Yu et al. (2017). Figure 1 provides an overview of the
study flow, adapted from the SAFE methodology. To
develop and evaluate algorithms for predicting AIS,
we employed the PheCAP R package, which
integrates medical codes and textual data as candidate
features in various classification methods. The SAFE
methodology allowed us to identify features closely
associated with AIS, where surrogate variables served
as “silver-standard labels” representing textbook
cases. These labels guided the selection of features for
algorithm training.
Figure 1: Study Flow Chart Simplified from SAFE (16).
3.2 AIS Data Mart Creation
Data were collected from the electronic medical
records (EMR) of the University of Kentucky
Healthcare System (UKHC), a large academic
medical centre with EMR data for over one million
patients since 2004. We screened structured EMR
data to identify potential AIS cases, including patients
with at least one ICD-9/-10 code specific to AIS
(M04.1, M04.8, M04.9), adult-onset Still’s disease
(M06.1 or 714.2), Behcet's disease (BD, 136.1 or
711.2x), cryopyrin-associated periodic syndromes
(CAPS, M04.2), or familial Mediterranean fever
(FMF, 277.31). To broaden our capture, we included
codes related to arthritis (714.2, 714.3, M06.9) and
National Drug Codes (NDCs) for AIS-related
medications such as anakinra, canakinumab, and
rilonacept. Patients under 18 at the time of diagnosis
or medication use were excluded. This preliminary
screening identified 273 patients for potential
inclusion in the AIS data mart.
3.3 Textual Data and Cohort
Refinement
We extracted narrative text data from multiple
clinical notes (e.g., outpatient, rheumatology,
discharge summaries) available in the EMR for each
patient. Only notes exceeding 500 characters were
used to ensure data quality. To refine our cohort
further, we included only patients with at least two
qualifying notes, resulting in a final dataset of 206
patients. Each patient was then classified as AIS or
non-AIS through manual chart review by an attending
rheumatologist, creating a set of gold-standard labels
for model training and validation.
HEALTHINF 2025 - 18th International Conference on Health Informatics
868

3.4 Feature Extraction and Codified
Data
A comprehensive set of structured codes and
unstructured data features was developed to define the
AIS phenotype. Our clinical expert, in collaboration
with SAFE and PheCAP developers, identified critical
AIS-related symptoms (e.g., “fever,” “rash”),
laboratory findings (e.g., “ferritin levels”), and
treatments (e.g., “IL-1 inhibitors”) based on clinical
experience. These terms were mapped to structured
EMR data sources such as ICD codes, CPT codes,
NDCs, and laboratory test identifiers (LOINC).
3.5 cNLP-Derived Features
We manually curated phenotype definitions for five
AIS subtypes (BD, CAPS, PFAPA, FMF, AOSD)
from publicly available sources (e.g., Medscape,
Mayo Clinic, MedlinePlus). Using the Unified
Medical Language System (UMLS), we identified
relevant clinical concepts and mapped them to unique
concept identifiers (CUIs). The Clinical Language
Annotation, Modelling, and Processing Toolkit
(CLAMP) software was then used to process 172,679
clinical notes, extracting only directly associated
concepts while excluding negated terms and family
history mentions. CLAMP’s rule-based and machine
learning components enabled us to develop a
customized pipeline for comprehensive extraction of
all relevant AIS concepts.
3.6 Model Development and Evaluation
Three supervised learning algorithms—adaptive
lasso penalized regression (ALASSO), support vector
machine (SVM), and random forest (RF)—were
adapted using the PheCAP pipeline to predict AIS
status. The dataset comprised 206 patient
observations and 199 variables, with 61 patients
labelled AIS-positive and 145 labelled non-AIS. To
evaluate performance, 40% of the data was reserved
for validation, while the remaining 60% was used for
training.
3.7 Surrogate Labelling and Feature
Selection
Our clinical expert identified key ICD and cNLP
features as surrogate “silver-standard” labels for the
SAFE process. These features included total counts
of AIS-related ICD codes (SICD) and cNLP-derived
mentions (SNLP), as well as a combined feature set
(SICDNLP = SICD + SNLP). Using penalized
logistic regression on these features, the SAFE
process selected 44 critical variables for final
algorithm training, aligning with expert choices.
3.8 Training and Validation
We trained the ALASSO, SVM, and RF models using
the 44 selected features, performing 200 training
iterations per model with randomized 70% data splits
for each iteration. Model performance was evaluated
on the training set through metrics such as the area
under the receiver operating characteristic (ROC)
curve (AUC), false positive rate (FPR), true positive
rate (TPR), positive predictive value (PPV), negative
predictive value (NPV), and F1 score. The validation
set was used for final model evaluation, with AUC,
sensitivity, specificity, PPV, and NPV calculated for
each algorithm.
4 RESULTS
4.1 Patient Characteristics
An initial pool of 273 potential AIS patients was
identified through medical claims data based on
relevant ICD-9/-10 codes and medication records. Of
these, 206 patients (75.46%) met the inclusion criteria,
each having at least two clinical notes of more than 500
characters in the EMR. The prevalence of confirmed
AIS within this final cohort was 29.6% (61 patients).
Demographic characteristics are summarized in Table
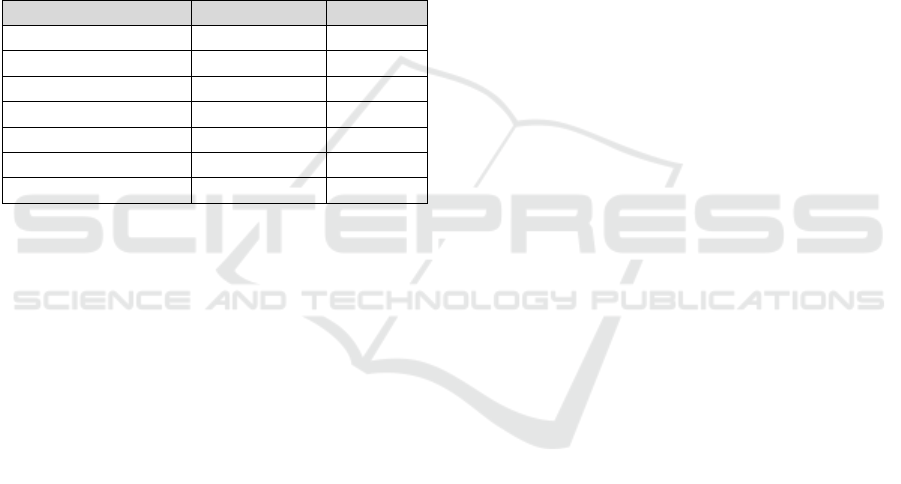
1. AIS patients were predominantly white (93.4%) and
female (63.9%), with a mean age of 40.8 years
(SD=13.9). The initial screening step involved using
IL-1 receptor antagonist medications as one criterion
for potential AIS cases, with anakinra being the most
commonly prescribed IL-1 receptor antagonist, used in
18.8% of AIS cases.
Table 1: Patient Characteristics from EMR.
(N, %) Overall Definite AIS Non-AIS
Total subjects 206 (100) 61 (29.6) 145 (70.4)
Age (Mean
years, SD)
40.7
(14.1) 40.8 (13.9) 40.7 (14.3)
Female 150 (72.8) 39 (63.9) 110 (75.9)
Race
-White 189 (91.7) 57 (93.4) 132 (91)
-Blac
k
14 (7.8) 3 (4.9) 11 (7.7)
-Asian 1 (0) 1 (1.6) 0 (0)
-Unreported 2 (1) 0 (0) 2 (1.4)
IL-1/IL-1R blocke
r
-Anakinra 18 (8.8) 9 (14.8) 9 (6.2)
-Rilonacept 2 (1) 2 (3.3) 0 (0)
-Canakinumab 5 (2.4) 4 (8.3) 1 (0.7)
Identifying an Autoinflammatory Syndrome Cohort Using Natural Language Processing with Electronic Medical Record Data
869
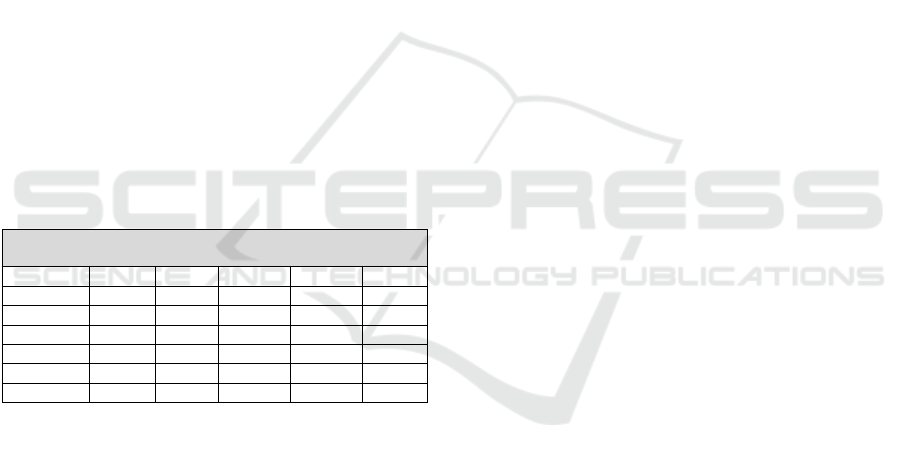
Treatment patterns within the AIS cohort are
presented in Table 2. Among AIS patients, IL-1/IL-
1R antagonists and anti-TNF medications were each
prescribed to 21.3% of patients. Glucocorticoids were
prescribed to 23% of AIS patients, while colchicine,
an anti-inflammatory medication frequently used in
autoinflammatory syndromes, was the most
prescribed medication, used by 32.8% of patients.
Immunosuppressant drugs were prescribed in 18% of
AIS cases, whereas NSAIDs were the least common
medication group, used by 3.3% of patients. Non-
biologic disease-modifying antirheumatic drugs
(nbDMARDs) were prescribed to 14.8% of the AIS
cohort, indicating moderate use of traditional
immunomodulatory therapies.
Table 2: Cohort treatment characteristics from EMR.
N (%) AIS Non-AIS
Anti-TNF 13 (21.3) 0 (0)
IL-1/IL-1R antagonis
t
13 (21.3) 1 (1)
Colchicine 20 (32.8) 1 (1)
Glucocorticoids 14 (23) 0 (0)
Immunosuppressan
t
11 (18) 0 (0)
nbDMARD 9 (14.8) 0 (0)
NSAIDs 2 (3.3) 0 (0)
4.2 Feature Extraction and Selection
for AIS Algorithms
Using a combination of structured ICD codes and
unstructured narrative data, our knowledge sources
produced 1,469 unique Unified Medical Language
System (UMLS) concepts as initial candidate
features. After applying a majority vote selection
process, 155 concepts met the threshold for inclusion,
of which 143 were found within clinical narratives.
To refine feature selection further, we applied the
SAFE methodology using penalized logistic
regression, which identified 44 key features highly
predictive of AIS. Notably, SAFE’s selection of these
44 features matched those identified by our clinical
expert, providing validation of the feature selection
process.
Among the final 44 features, only 10 had a
statistically significant impact on model performance,
including four ICD codes and six UMLS-derived
concepts. The ICD codes included:
• Rheumatoid arthritis (714.2, 714.3): These
codes, although traditionally associated with
autoimmune conditions, were predictive in the
AIS model, possibly due to overlapping
inflammatory symptoms.
• Behcet’s disease (M35.2): This code directly
aligns with AIS manifestations and contributed
substantially to the model.
• Juvenile chronic polyarthritis (M06.1):
Interestingly, this code showed a negative
association with AIS diagnosis, suggesting it
may serve as a distinguishing factor for non-AIS
cases within the algorithm.
The six UMLS-derived concepts that enhanced
model prediction included clinical symptoms,
specific syndromes, and treatments:
• Symptoms: “Periodic fever” (C0015974) and
“oral lesions” (C0149744) were among the
selected features. Though common across other
conditions, these symptoms are relevant to AIS
and were consistently identified in clinical
narratives.
• Specific Syndromes: “Hypopyon” (C0020641), a
symptom of eye inflammation frequently seen in
Behcet’s disease, was selected due to its
specificity. “Muckle-Wells syndrome”
(C0268390), a subtype of cryopyrin-associated
periodic syndromes (CAPS), had a strong
association with AIS, though CAPS codes were
not predictive in themselves. Finally,
“macrophage activation syndrome” (C1096155),
a severe complication of systemic autoimmune
diseases, also showed positive predictive value
for AIS.
• Treatment: Colchicine was uniquely impactful,
not as a general medication, but specifically as a
coded therapeutic procedure for colchicine
treatment (C0742540), suggesting that recorded
instances of colchicine intervention are more
predictive of AIS status than mere prescription
records.
A full list of the selected features and their
classification roles within the three final models is
available in the Appendix.
4.3 Model Performance and Validation
Three machine learning algorithms—ALASSO,
SVM, and RF—were trained using the 44 selected
features to classify AIS. Each model’s performance
was initially evaluated on a training set and then on a
validation set, with results summarized below.
The ALASSO model demonstrated a high AUC
of 0.996 on the training set, showing strong
sensitivity and NPV. However, when applied to the
validation set, the AUC dropped slightly to 0.94, with
sensitivity also reduced, although NPV remained
high. Importantly, PPV showed consistent
HEALTHINF 2025 - 18th International Conference on Health Informatics
870
performance across training splits, suggesting that the
model’s predictive power is stable but may benefit
from further refinement to improve sensitivity.
Both SVM and RF models exhibited perfect
classification performance on the training data (AUC
= 1.0). On the validation set, these models
outperformed the ALASSO model, with AUC values
of 0.954 for SVM and 0.948 for RF. These models
also showed an increase in metrics such as PPV and
TPR when compared to ALASSO, except for the
FPR, which remained steady across all models. This
consistency in FPR indicates reliable specificity
across algorithms, though further testing is necessary
to assess their robustness in larger datasets.
Using these models to predict the probability of
AIS phenotype among patients, the majority were
classified with either a high likelihood (>90%) or low
likelihood (<10%) of AIS. Table 3 presents a
comparative overview of evaluation metrics,
including TPR, PPV, NPV, and F1 scores at fixed
FPRs of 0 and 0.195. These metrics illustrate the
models’ abilities to maintain strong predictive
performance with consistent precision and recall,
especially at a controlled FPR level, highlighting the
potential for these algorithms in accurately
identifying AIS cases.
Table 3: Comparison of evaluation metrics at fixed FPRs.
Comparisons of TPR, PPV, NPV, and F1 scores at fixed
FPR
FPR TPR PPV NPV F1
ALASSO 0 0.362 1 0.791 0.531
0.195 1 0.679 1 0.809
SVM 0 0.486 1 0.825 0.655
0.195 1 0.679 1 0.809
RF 0 0.486 1 0.825 0.655
0.195 1 0.679 1 0.809
5 DISCUSSIONS
The integration of cNLP was instrumental in the
development of the AIS phenotype algorithm, enabling
the incorporation of rich clinical data unavailable
through structured coding alone. Codified data, such as
ICD codes, often lack the granularity required for rare
conditions like AIS and are subject to inconsistent
application. cNLP offers a solution by extracting
detailed clinical information from unstructured
narrative text, allowing for a deeper understanding of
complex conditions. This study demonstrated the
potential of cNLP to identify episodic flare-ups and
atypical presentations of AIS, highlighting its value for
rare disease phenotyping (Ramirez, 2012; Liao, 2017;
Ananthakrishnan, 2013; Liao, 2015).
Despite its benefits, cNLP applications are not
without challenges. Linguistic ambiguities, variations
in clinical documentation, and the use of non-
standard terminology can reduce the precision of
cNLP-derived features. Nonetheless, unstructured
clinical notes provide a wealth of information not
captured in traditional claims-based research (Lenert,
2020a; Lenert, 2020b). This is particularly important
for AIS, where the distinctive characteristics of the
disease, such as symptom variability and treatment
patterns, may not be adequately represented by
structured codes. Traditional approaches relying
solely on claims data fail to capture these subtleties,
underscoring the necessity of incorporating cNLP
into phenotyping workflows.
The rarity of AIS introduces unique challenges in
algorithm development. With a low prevalence in the
population, achieving a high PPV often results in
missed cases due to overly stringent criteria. By
balancing PPV and NPV, this study ensured
comprehensive case capture while maintaining model
accuracy. The combination of cNLP and machine
learning provided an adaptable framework to
optimize phenotyping for AIS, adapting proven
protocols for rare diseases to our unique dataset (Liao,
2017; Ananthakrishnan, 2013; Zheng, 2014).
The SAFE method played a crucial role in feature
selection, identifying 44 predictive features from an
initial pool of 1,469 candidate variables. SAFE’s
alignment with features selected by clinical experts
validates its utility in streamlining the feature
selection process. Importantly, SAFE excluded
generalized terms, such as “very high” or “very rare,”
which lack clinical specificity, resulting in a more
refined and meaningful feature set. This ability to
automate feature refinement while maintaining
alignment with expert curation suggests that SAFE
has significant potential for phenotyping rare diseases
with minimal human intervention. Future studies
could explore how SAFE might be fine-tuned to
further reduce reliance on expert oversight without
compromising the accuracy of selected features.
Another significant observation was the
comparable performance of the three supervised
learning algorithms—ALASSO, SVM, and RF—
using the same 44 features. The slight differences in
results suggest that the quality of feature selection has
a greater impact on model performance than the
specific algorithm employed. This reinforces the
critical role of feature selection in phenotyping rare
diseases, where the selection of informative features
is often limited by small sample sizes.
The study also sheds light on the clinical validity
of certain features through administrative codes. For
Identifying an Autoinflammatory Syndrome Cohort Using Natural Language Processing with Electronic Medical Record Data
871

example, the inclusion of ICD codes for rheumatoid
arthritis and Behcet’s disease highlights overlapping
inflammatory pathways with AIS, while the negative
association of juvenile chronic polyarthritis (M06.1)
suggests it may serve as a distinguishing feature for
non-AIS cases. Similarly, UMLS-derived concepts
such as “hypopyon” and “macrophage activation
syndrome” contributed strongly to the model,
reflecting the complexity of AIS and its associations
with other inflammatory syndromes. Interestingly,
“colchicine treatment” was predictive of AIS,
emphasizing the importance of capturing therapeutic
interventions rather than merely listing prescribed
medications.
The inclusion of multiple supervised learning
algorithms allowed for robust model comparison.
ALASSO performed well in training but showed
slightly reduced sensitivity on validation, while SVM
and RF models demonstrated stronger generalization
with validation AUCs of 0.954 and 0.948, respectively.
The consistency of false positive rates (FPR) across
models underscores their reliability in distinguishing
AIS from non-AIS cases. These findings highlight the
value of combining machine learning with expert-
curated and NLP-derived features to create adaptable,
high-performing algorithms.
Beyond its methodological contributions, this
study has implications for clinical and translational
research. By providing a scalable framework for AIS
identification, this work can facilitate the creation of
larger, well-characterized cohorts for epidemiological
and interventional studies. Accurate AIS phenotyping
may also support precision medicine initiatives by
enabling targeted analyses of treatment outcomes and
disease progression in diverse patient populations.
However, achieving widespread adoption of such
algorithms requires addressing barriers to
implementation. Portability remains a major concern,
as differences in EMR systems, documentation
practices, and linguistic conventions can limit
reproducibility. External validation across multiple
institutions with diverse populations is essential to
ensure that these algorithms are generalizable and
robust. Additionally, collaboration with clinicians,
especially paediatric rheumatologists, could expand
the algorithm’s applicability to younger populations,
addressing the unmet need for AIS phenotyping in
paediatric patients.
This study also emphasizes the importance of
multidisciplinary collaboration in phenotyping
research. The integration of clinical expertise,
computational methods, and cNLP tools exemplifies
the potential of interdisciplinary approaches to
overcome the limitations of traditional claims-based
methodologies. By continuing to refine these
methods and expand their applications, this
framework has the potential to transform rare disease
research and improve patient outcomes.
This study had several limitations. First, the
relatively small cohort size (206 patients, with 61
confirmed AIS cases) increases the risk of overfitting
and limits generalizability. Future studies should
validate these findings using larger, multicentre
datasets. Second, excluding patients under 18
potentially omits paediatric AIS cases, which may
differ from adult phenotypes and restricts the
algorithm's broader applicability. Third, variability in
educational resources for the five AIS subtypes may
have biased feature selection. While majority voting
reduced this issue, certain subtypes may still be
under- or overrepresented, warranting more balanced
data sources in future work. Finally, reliance on
cNLP-derived features poses portability challenges,
as differences in EMR systems and documentation
practices may affect reproducibility. External
validation across diverse EMR platforms will be
essential to ensure robustness and generalizability.
ACKNOWLEDGEMENTS
This study was partially supported by the VERITY
Pilot & Feasibility Research Award (principal
investigator: A.L.; coinvestigator: S.K.) from the
Brigham and Women's Hospital and NIH-NIAMS
(P30-AR-072577). The content is solely the
responsibility of the authors and does not necessarily
represent the official views of the NIH.
REFERENCES
Ananthakrishnan, A. N., Cai, T., Savova, G., et al. (2013).
Improving case definition of Crohn's disease and
ulcerative colitis in electronic medical records using
natural language processing: A novel informatics
approach. Inflammatory Bowel Diseases, 19, 1411–
1420.
Barbhaiya, M., Feldman, C. H., Guan, H., et al. (2017).
Race/ethnicity and cardiovascular events among
patients with systemic lupus erythematosus. Arthritis &
Rheumatology, 69, 1823–1831.
Brownstein, J. S., Murphy, S. N., Goldfine, A. B., et al.
(2010). Rapid identification of myocardial infarction risk
associated with diabetes medications using electronic
medical records. Diabetes Care, 33, 526–531.
Desai, R. J., & Solomon, D. H. (2017). Use of large
healthcare databases for rheumatology clinical research.
Current Opinion in Rheumatology, 29, 138–143.
HEALTHINF 2025 - 18th International Conference on Health Informatics
872

Feldman, C. H., Hiraki, L. T., Liu, J., et al. (2013).
Epidemiology and sociodemographics of systemic
lupus erythematosus and lupus nephritis among US
adults with Medicaid coverage, 2000–2004. Arthritis &
Rheumatism, 65, 753–763.
Feldman, C. H., Hiraki, L. T., Winkelmayer, W. C., et al.
(2015). Serious infections among adult Medicaid
beneficiaries with systemic lupus erythematosus and
lupus nephritis. Arthritis & Rheumatology, 67, 1577–
1585.
Hak, A. E., Karlson, E. W., Feskanich, D., Stampfer, M. J.,
& Costenbader, K. H. (2009). Systemic lupus
erythematosus and the risk of cardiovascular disease:
Results from the nurses' health study. Arthritis &
Rheumatism, 61, 1396–1402.
Hintenberger, R., Falkinger, A., Danninger, K., &
Pieringer, H. (2018). Cardiovascular disease in patients
with autoinflammatory syndromes. Rheumatology
International, 38, 37–50.
Kim, S. C., Solomon, D. H., Rogers, J. R., et al. (2017).
Cardiovascular safety of tocilizumab versus tumor
necrosis factor inhibitors in patients with rheumatoid
arthritis: A multi-database cohort study. Arthritis &
Rheumatology, 69, 1154–1164.
Kim, S. Y., Servi, A., Polinski, J. M., et al. (2011). Validation
of rheumatoid arthritis diagnoses in health care utilization
data. Arthritis Research & Therapy, 13, R32.
Lenert, A., Oh, G., Ombrello, M. J., & Kim, S. (2020a).
Clinical characteristics and comorbidities in adult-onset
Still’s disease using a large US administrative claims
database. Rheumatology (Oxford).
Lenert, A., Russell, M. J., Segerstrom, S., & Kim, S.
(2020b). Accuracy of US administrative claims codes
for the diagnosis of autoinflammatory syndromes.
Journal of Clinical Rheumatology.
Liao, K. P., Ananthakrishnan, A. N., Kumar, V., et al.
(2015). Methods to develop an electronic medical
record phenotype algorithm to compare the risk of
coronary artery disease across 3 chronic disease
cohorts. PLoS One, 10, e0136651.
Liao, K. P., Cai, T., Gainer, V., et al. (2010). Electronic
medical records for discovery research in rheumatoid
arthritis. Arthritis Care & Research (Hoboken), 62,
1120–1127.
Liao, K. P., Cai, T., Savova, G. K., et al. (2015).
Development of phenotype algorithms using electronic
medical records and incorporating natural language
processing. BMJ, 350, h1885.
Liao, K. P., Diogo, D., Cui, J., et al. (2014). Association
between low density lipoprotein and rheumatoid
arthritis genetic factors with low density lipoprotein
levels in rheumatoid arthritis and non-rheumatoid
arthritis controls. Annals of the Rheumatic Diseases,
73, 1170–1175.
Liao, K. P., Sparks, J. A., Hejblum, B. P., et al. (2017).
Phenome-wide association study of autoantibodies to
citrullinated and noncitrullinated epitopes in
rheumatoid arthritis. Arthritis & Rheumatology, 69,
742–749.
McGonagle, D., & McDermott, M. F. (2006). A proposed
classification of the immunological diseases. PLoS
Medicine, 3, 12428.
Ramirez, A. H., Shi, Y., Schildcrout, J. S., et al. (2012).
Predicting warfarin dosage in European-Americans and
African-Americans using DNA samples linked to an
electronic health record. Pharmacogenomics, 13, 407–
418.
Ridker, P. M. (2016). From C-reactive protein to
interleukin-6 to interleukin-1: Moving upstream to
identify novel targets for atheroprotection. Circulation
Research, 118, 145–156.
Yu, S., Chakrabortty, A., Liao, K. P., Cai, T.,
Ananthakrishnan, A. N., Gainer, V. S., et al. (2017).
Surrogate-assisted feature extraction for high-
throughput phenotyping. Journal of the American
Medical Informatics Association, 24(e1), e143–e149.
Zhang, Y., Cai, T., Yu, S., Cho, K., Hong, C., Sun, J., et al.
(2019). High-throughput phenotyping with electronic
medical record data using a common semi-supervised
approach (PheCAP). Nature Protocols, 14(12), 3426–
3444.
Zheng, C., Rashid, N., Wu, Y. L., Koblick, R., Lin, A. T.,
Levy, G. D., & Cheetham, T. C. (2014). Using natural
language processing and machine learning to identify
gout flares from electronic clinical notes. Arthritis Care
& Research, 66(11), 1740–1748.
APPENDIX
The below table lists the features which were used in
all three of the final training algorithms along with the
gold-standard labels. Features with non-zero beta
coefficients for the ALASSO model are highlighted
in bold.
AIS features extracted from SAFE
Claims
code
M06.1, 714.20, 714.30, 136.1, M35.2, M04.2,
277.31, M04.1, M04.8, M04.9
UMLS
features
(CUI:
Concept
Names)
C0040423: tonsillectomy, C003864: anakinra,
C0042164: uveitis, C0151281: genital ulcers,
C0009262: colchicine, C0031350: pharyngitis,
C0009763: conjunctivitis, C0031154: peritonitis,
C0031046: pericarditis, C0152031: swollen joints,
C0149745: oral ulcers, C0037198: sinus
thrombosis, C0010592: cyclosporine, C1609165:
tocilizumab, C0027059: myocarditis, C0015974:
periodic fever, C0149744: oral lesions, C2718773:
canakinumab, C0031069: familial Mediterranean
fever, C0001416: adenitis, C0152026: retinal
vasculitis, C2343589: rilonacept, C0277799:
episodic fever, C0038363: aphthous stomatitis,
C0343068: familial cold autoinflammatory
syndrome, C1510431: superficial
thrombophlebitis, C3161802: pathergy test,
C0018784: sensorineural deafness, C1096155:
macrophage activation syndrome, C0268390:
muckle wells syndrome, C0847014: fever rash,
C0020641: hypopyon, C0424781: fever spikes,
C0742540: colchicine treatment
Identifying an Autoinflammatory Syndrome Cohort Using Natural Language Processing with Electronic Medical Record Data
873
