
Medical Chatbot for Disease Prediction Using Machine Learning and
Symptom Analysis
Oltean Anisia Veronica
a
, Ioan Daniel Pop
b
and Adriana Mihaela Coroiu
c
“Babes-Bolyai” University, Department of Computer Science, 400084, Cluj-Napoca, Romania
Keywords:
Disease Prediction, Logistic Regression, Random Forests, Decision Trees, Naive Bayes, Multilayer
Perceptron.
Abstract:
This paper emphasizes the transformational role of artificial intelligence in the medical field by studying not
only various machine learning algorithms used for symptoms-based disease prediction, but also methods used
in conversational artificial intelligence. At its core, the research was carried out as a first step in the devel-
opment of a medical chatbot that allows patients to receive diagnosis and advice related to various diseases
and their possible treatments. In our paper, various machine learning algorithms were compared for predict-
ing diseases based on symptoms, such as Logistic Regression, Random Forests, Decision Trees, Naive Bayes
and Multilayer Perceptron, which were evaluated on multiple datasets. Given the lack of publicly available
datasets for such a task, a final dataset was generated, achieving satisfactory accuracy values of approximately
80%.
1 INTRODUCTION
Health is one of the most important things in a per-
son’s life. Studies on improving health date back
thousands of years. Considering the advance of tech-
nology in the last decades, it is not at all surprising
that various studies have been carried out regarding
its integration into the medical system. Chatbots are
among the technological tools most used in the med-
ical field. Chatbots are used in various fields, among
the most well-known being health, customer relations
and finance services, with the main advantage of im-
proving interaction with customers through less hu-
man intervention (an example would be virtual agents
that assist in placing an order and are available 24/7,
without the need to call a call center). They have be-
come increasingly popular in recent years due to ad-
vancements in artificial intelligence and natural lan-
guage processing, they are proving increased skills
and performance in understanding users and provid-
ing coherent answers. As the source (Siddique and
Chow, 2021) mentions, in the field of healthcare, chat-
bots have a huge development potential, because they
can improve the patient experience through remote
a
https://orcid.org/0009-0003-7706-9583
b
https://orcid.org/0000-0002-3740-6579
c
https://orcid.org/0000-0001-5275-3432
monitoring, being able to provide quick and timely
responses. Access to health is essential for everyone,
but scheduling a medical consultation is not always
a viable solution due to long waiting times and high
costs, a medical chatbot can be the solution to such
problems. For such a conversational agent to be effec-
tive, it needs to process and analyze user queries, ex-
tract relevant information such as symptoms and pro-
vide a diagnosis as accurate as possible, accompanied
by any advice that should be followed (treatments,
scheduling a consultation or even an emergency visit
to the doctor). It is important that the chatbot has a
natural language dialogue with the patient and pro-
vides personalized answers, depending on the condi-
tion and symptoms identified. In essence, chatbots
represent valuable tools in medicine, whose purpose
is not to replace medical staff, but rather to highlight
an interdisciplinary approach between technology and
specialists to benefit patients (Altamimi et al., 2023).
Considering all of this, our paper aims to highlight
the role of artificial intelligence in medicine, address-
ing two main topics, namely the use of algorithms to
predict diseases based on symptoms and the develop-
ment of a medical chatbot to facilitate dialogue with
patients through natural language. Therefore, the cre-
ated chatbot has as main objectives:
• recognizing symptoms from user phrases
• collection of associated symptoms, simulating the
600
Veronica, O. A., Pop, I. D. and Coroiu, A. M.
Medical Chatbot for Disease Prediction Using Machine Learning and Symptom Analysis.
DOI: 10.5220/0013357900003928
In Proceedings of the 20th International Conference on Evaluation of Novel Approaches to Software Engineering (ENASE 2025), pages 600-607
ISBN: 978-989-758-742-9; ISSN: 2184-4895
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

dialogue between doctor and patient
• providing a diagnosis for the symptoms found (as-
sumes training the classification model)
• to provide explanations and possible treatments
for the diseases identified
2 LITERATURE REVIEW
In light of the importance of the field, there are a va-
riety of studies that address this topic, in the follow-
ing we will analyze the studies that we considered the
most relevant.
The first analyzed case study (Tode et al., 2021),
the chatbot CureBot proposes a relatively simple ar-
chitecture in building the bot, the preprocessing of the
message from the user is done with the Natural Lan-
guage Toolkit (NLTK) library to ignore the punctua-
tion. Phrases/sentences are turned into tokens, after
which the bag of words technique is applied to en-
code words into numerical data. The neural network
model is created using the Keras framework (sequen-
tial, with 2 hidden starts), softmax activation function
and SGD (stochastic gradient descent) optimization
for sentence intent classification. The training takes
place based on a file in .json format called ’intents’
that contains the tag, followed by a series of phrases
for matching (pattern matching) and a series of sen-
tences to generate the response. Although the source
of the data or the accuracy of the model is not men-
tioned, the author mentions that the model is built to
identify the presence or absence of Covid-19 infec-
tion based on user inputted symptoms. In a similar
way, the HELPI chatbot (Karuna et al., 2023) is built,
which uses the data set ”The Disease Symptom Pre-
diction Dataset”, available on Kaggle (approx. 5000
rows) with 133 columns (the first 132 represent the
symptoms, and the last one is the disease), there be-
ing 42 possible diseases (diagnoses) in total. The pre-
diction of the diagnosis takes place through decision
trees, using the CART (Classification and Regression
Trees) algorithm, but even in this case no metrics are
mentioned regarding the performance of the model.
The next model analyzed is Diabot (Bali et al.,
2019), a chatbot built specifically for diabetes pre-
diction based on user-input symptoms. On the back-
end, the RASA framework is used, and the classifica-
tion problem (presence/absence of the disease) is the
result of an ensemble learning approach.The dataset
used contains training on 768 women from a popu-
lation in Phoenix, Arizona, USA, of which 258 had
diabetes and the remaining 500 did not. There are 9
attributes in total (8 represent the factors considered in
disease prediction, the last one being the target vari-
able – 0 or 1). When training the model, the result of
6 models (Multinomial Na
¨
ıve Bayes, Decision Trees,
Random Forest, k-NN, Logistic Regression and Gra-
dient Boost) are combined, each of them being trained
individually. The final decision is made by voting, us-
ing the majority voting algorithm – of the two possi-
ble classes (0 or 1), the one predicted by most of the
models wins. The proportion of the data set used for
training and testing is 80 – 20%, with the accuracy of
the model reaching 82%.
The Kiwi chatbot (Chakraborty et al., 2022) is
a purpose-built model to answer questions/queries
about the Covid-19 infection using information from
a dataset built by the author. Similar to the first ana-
lyzed model (Tode et al., 2021), the data is organized
in a JSON file containing several tags representing the
categories of information in which the user’s message
will have to be classified. Each category contains
several ’patterns’, phrases that describe examples of
possible queries that can come from the user. The
model will choose the most appropriate response from
a set of predefined responses. Before building the
model, language processing techniques are applied
to pre-process the text such as punctuation ignoring
and lemmatization, and the bag of words technique
is used to map the words/phrases as numbers. The
built model is based on a neural network with 3 layers,
among which the hidden layer has the ReLu activation
function, while the output layer has softmax. Categor-
ical Crossentropy or Adam were used as optimizers
for the model. When testing the model, the author
uses an improved model with encoder-decoder archi-
tecture (assumes the addition of LSTM - Long Short
Term Memory layers), but the configuration is not dis-
closed. The accuracy of the model reaches 94% and
is compared to other possible methods tried (recurrent
neural networks RNN or decision trees), but this is the
variant with the best results.
The following analyzed article (Vasileiou and Ma-
glogiannis, 2022) proposes the development of an in-
telligent system based on dialogue with application
in telemedicine. Thus, the authors propose a chatbot
model developed with the DialogFlow conversational
AI platform. The NLP component of the platform
deals with analyzing the text, establishing the user’s
intention (intent classification) and also identifying
keywords. The system response is either predefined
(a response is chosen from the training set introduced
in the platform) or from an ML engine (ML Engine
– used for diagnostics). The Accuracy of the model
reaches 98.3%. The second model, Heart Disease,
uses as data set the Cleveland Heart Dataset (UCI)
– composed of 303 medical records with 14 attributes
Medical Chatbot for Disease Prediction Using Machine Learning and Symptom Analysis
601

that are taken into account when predicting the dis-
ease). The data set split is 33% testing and 67% train-
ing, here using several classification models based on
the sklearn library – logistic regression, SVC (sup-
port vector classifier), Gaussian Naive Bayes Classi-
fier, Decision Tree Classifier (decision trees) and Ran-
dom Forest. The best performing model was the lo-
gistic regression model, with 82% accuracy.
In the paper (Polignano et al., 2020), the authors
propose a personal medical assistant called HealthAs-
sistantBot (HOB), specialized for the Italian lan-
guage. The interaction with the conversational agent
takes place through the Telegram platform, the built
system having 2 main tasks: Intent Recognition and
Entity Recognition. Classification algorithms such
as Naive Bayes, Logistic Regression, Decision Tree
Forest and a Multilayer Perceptron Network were use
to create the model. The performance of the model
was tested considering metrics such as accuracy and
F1 score, and it was found that the Naive Bayes
model performed best for k between 1000 and 2500,
with values for accuracy and F1 score equal to 0.942
(k=1000) and 0.87 ( k=2500).
In the paper (Shedthi B et al., 2023), the authors
propose the development of a website where users
can communicate different aspects related to health,
as well as with an integrated chatbot that has the
task of identifying the user’s symptoms and provid-
ing a diagnosis based on machine learning algorithms.
It is considered that a minimum of 3 symptoms are
needed for the prediction of the disease to occur. The
dataset used is available on Kaggle (133 columns and
41 symptoms), where values of 1 (symptom present)
or 0 (symptom absent) can appear on each row. All
the algorithms used in the work provide over 90% ac-
curacy. In Table 1 it can be seen the result for each
algorithm. In the table below, the following abbrevi-
ations were used: Support Vector Machine as SVM,
Random Forest as RF, K-Nearest Neighbors as KNN,
Bayesian Network as BN and Logistic Regression as
LG.
Table 1: The performances of the models presented in the
paper (Shedthi B et al., 2023).
ACC Precision Recall F1 Score
SVM 0.9079 0.91 0.91 0.90
RF 0.9737 0.98 0.97 0.97
KNN 0.9079 0.89 0.91 0.89
NB 0.9605 0.98 0.96 0.97
LR 0.9474 0.97 0.95 0.94
The last work, (Babu and Boddu, 2024), uses a
pre-trained BERT (Bidirectional Encoder Represen-
tations from Transformers) model used to obtain con-
textualized embeddings that are then used in tasks
such as entity recognition and intent classification.
Fine-tuning is done on a set to generate responses
of approximately 11,000 question-answer pairs col-
lected from various sources (MIMIC-III, PubMed,
BioASQ, etc.). The model achieves high accuracy
(98%) for processing medical queries, but despite the
obtained metrics, the authors highlight some prob-
lems such as the high time required for training and
the possible decrease in model efficiency if there is
not enough training data for some medical cases.
3 THEORETICAL BACKGROUND
Symptom-based disease diagnosis, a central topic
in our paper, is an application of supervised learn-
ing–namely multi-class classification. The input vari-
ables (features) are represented by a list of symptoms
that will need to be processed into numerical form in
order to be processed by the AI-based classification
algorithms. The target variable y is a discrete variable
and it can take values from the set of all diagnoses
(diseases), the number of which varies depending on
the data set used. Formally, the diagnosis problem
involves finding a model that, for an input data set
(x
1
, x
2
, ..., x
m
) where x
i
represents a symptom and m is
the total number of symptoms, assigns a label y rep-
resenting the diagnosis (disease). y is part of a finite
set of diagnoses D with card(D) = n, so y can take any
value from the set of n possible diagnoses. An ex-
ample for D could be {appendicitis, pneumonia, flu,
indigestion}, where there are n=4 possible diagnoses,
and for x=(cough, fever, chest pain) a possible list
of symptoms from the set {cough, abdominal pain,
fever, chest pain, constipation, flatulence} etc.
3.1 Methods
In this work, we used five algorithms for disease
prediction based on symptoms: Logistic Regression,
Gaussian Naive Bayes, Random Forest, Decision Tree
and MultiLayer Perceptron. All these algorithms
were used in the multiclass classification task.
Logistic Regression (LR) is a statistical model
used for binary classification, predicting the probabil-
ity that a given input belongs to one of two classes.
Unlike linear regression, which predicts continuous
values, logistic regression uses the logistic function
(sigmoid) to output probabilities between 0 and 1 (Za-
bor et al., 2022). The model estimates the param-
eters (weights) by maximizing the likelihood of the
observed data. It is widely used in machine learning
and statistics for problems like spam detection, medi-
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
602

cal diagnosis, and credit scoring, where the goal is to
classify inputs into two distinct categories based on
various features (Zabor et al., 2022).
A variation of the Naive Bayes classifier that
makes the assumption that the features have a nor-
mal (Gaussian) distribution is called Gaussian Naive
Bayes (GNB). Based on the idea that features are in-
dependent of one another, it computes the probability
of each class given the input features. This method
is known as Bayes’ Theorem. The likelihood of the
features is represented by a Gaussian distribution for
Gaussian Naive Bayes, which is determined by its
mean and variance. It offers simplicity and efficiency
and is especially useful for classification tasks where
the input data is continuous, like medical diagnosis
spam detection and text classification (Reddy et al.,
2022).
Random Forest (RF) is an ensemble learning
method used for both classification and regression
tasks. It operates by constructing multiple decision
trees during training and combining their outputs to
improve accuracy and reduce overfitting. Each tree
is built using a random subset of the data, and each
node in a tree splits on the best feature from a random
subset of features. The final prediction is typically
made by averaging the predictions of all trees (for
regression) or by majority voting (for classification).
Random Forests are robust, handle large datasets well,
and are resistant to noise and overfitting (Probst et al.,
2019).
A Decision Tree (DT) is a supervised machine
learning algorithm used for classification and regres-
sion tasks. It models decisions as a tree-like structure,
where each internal node represents a test on a fea-
ture, each branch represents the outcome of the test,
and each leaf node represents a final decision or pre-
diction. The tree is built by recursively splitting the
data into subsets based on the feature that provides the
highest information gain or the lowest impurity. Deci-
sion Trees are intuitive, easy to visualize, and handle
both numerical and categorical data, though they can
be prone to overfitting without pruning (Costa and Pe-
dreira, 2023).
A Multilayer Perceptron (MLP) is a type of arti-
ficial neural network composed of multiple layers of
nodes: an input layer, one or more hidden layers, and
an output layer. Each node (neuron) in the network,
except for the input layer, applies a nonlinear activa-
tion function to its inputs and passes the result to the
next layer. MLPs are trained using backpropagation
to minimize a loss function, adjusting the weights be-
tween neurons. They are capable of learning complex
patterns and are commonly used for tasks like classi-
fication, regression, and pattern recognition, but can
be computationally expensive (Almeida, 2020).
All five models have been tested, to see which one
best fits our problem.
4 DATASET
The collection of data sets has been a major impedi-
ment precisely because of their lack, as there are few
publicly available data sets. Thus, when establish-
ing the classification algorithms, we considered sev-
eral data sets collected from various platforms such
as Kaggle, HuggingFace or GitHub. The examples
described in this section include the analysis of the
collected data sets, and then the necessary process-
ing on them (elimination of duplicates, encoding of
symptoms in numerical form).
The first dataset used is (KaggleDataset) (Patil,
2024) and consists of a total of 132 symptoms and 42
possible diseases in a csv file. In total, there are 4920
rows, each simulating an example patient represented
by a list of symptoms and the associated disease. At
first glance, the distribution of patients per disease is
balanced – each disease has exactly 120 patient ex-
amples associated with it, but upon further analysis,
following the elimination of duplicate rows, the num-
ber of examples decreases from 4920 to 348, losing
at the same time and balancing the number of patients
per disease.
The second dataset used (LargeDatasetHF) (Anh,
2024) contains information on 392 diseases, in total
there are up to 892 distinct symptoms in the dataset.
Here, however, for each disease there is only one ex-
ample with associated symptoms (there is 1 sample
per class), and to test the accuracy of the classifica-
tion models we formed a test case with 83 diseases.
The next dataset analyzed is one of the datasets
synthetically generated by the authors of the paper
(Yuan and Yu, 2024) (MedlinePlus) available at (Yuan
and Yu, 2021). The set contains 893 diseases and
1556 symptoms in total and is similar to that of (Anh,
2024) in that there is only one example per disease.
For the final project, we manually selected 38 dis-
eases from the data obtained by the authors of the
paper (Yuan and Yu, 2024) from the other data files
available in the public Github repository (Symcat),
they being similar to those used in the paper (Polig-
nano et al., 2020) which makes possible similarity
of performance, where a similar data set adapted to
the Italian language is used. And here, in the case
of maintaining the large dimensions of the number
of diseases and symptoms, the accuracy of the mod-
els drops to approximately 60%, a phenomenon also
noted in the work described. For this reason we came
Medical Chatbot for Disease Prediction Using Machine Learning and Symptom Analysis
603
to the decision to reduce the data set (Symcat38), the
results improved.
An advantage of this generated data set is the oc-
currence of prevalence for each symptom associated
with the diseases, which makes it easier to generate
patients. The generation of a ”patient” consists in se-
lecting some symptoms from a list associated with
each disease in order to obtain several ”examples”
of various symptoms for a certain diagnosis. When
generating the patients, we chose to eliminate dupli-
cates in order not to have data from the training set
in the test set. However, this choice has the disadvan-
tage of some classes (diseases) that are represented
by fewer examples in training, these diseases being
predicted with a low probability in testing. Applying
tutor methods to a dataset that contains 100% real pa-
tients, could reduce the accuracy of the system a little,
but certainly the adaptability of the system to real in-
puts would increase.
5 EXPERIMENTAL RESULTS
5.1 Encoding Symptoms
5.1.1 One-Hot Encoding
For all datasets used, a technique described in
(Hapke et al., 2019) called One-Hot, adapted for
symptom encoding, was used: if there are a to-
tal of M symptoms in a dictionary D, a list l =
(symptom 1, symptom 2, ..., symptom N) of symp-
toms will be represented as a vector of size M which
will have values of 1 only for indices in dictionary
D of symptoms in list l, and 0 otherwise. This tech-
nique is useful when working with categorical vari-
ables in machine learning models, since many algo-
rithms only work with numeric data, and categorical
variables must be converted to numbers to be pro-
cessed correctly.
5.1.2 Vector Embeddings
Since if the number of symptoms in an example is
low (3 or 4) this representation can lead to a sparse
array, we also tried transforming a list of symptoms
into a vector of real numbers of size 200 using word
embeddings (the vector representations) from the pa-
per (Zhang et al., 2019). Since a symptom can consist
of several words, for the representation we chose to
do the arithmetic mean of the vectors for each word.
For example, for ”sore throat” there are embeddings
for the words ”sore” and ”throat”, denoting by em(s)
the encoding vector obtained for the symptom (word)
s, then em(”sore throat”) we calculated it according to
the formula
em(”sore”)+ em(”throat”)
2
(1)
where dividing by 2 the resulting vector means divid-
ing by 2 each component of the vector. However, this
method gives very poor results for some algorithms
(see Table 2). LargeDatasetHF represents the origi-
nal dataset (where each disease has only one patient),
and LargeDatasetHFAugumented represents the same
dataset only that we generated patients. The compar-
ison between the two data sets highlights that large
data sets cannot achieve high accuracy results com-
pared to using other small data sets. When we talk
about the performance of the models, we are not only
referring to accuracy, considering the fact that we are
solving a classification problem, for the performance
evaluation, besides accuracy, we decided to use met-
rics such as precision, recall and F1-score. In Table 2
LR
Table 2: Prediction using Word Embeddings.
Kaggle LargeDatasetHF Augumented
LR 0.98 0.78 0.65
RF 0.98 0.79 0.61
DT 0.63 0.10 0.21
MLPC 1.0 0.85 0.65
GNB 0.95 0.01 0.51
5.2 Results and Discussion
For the classification of symptoms in diseases, we
used 5 algorithms from the scikit-learn library (Pe-
dregosa et al., 2011), namely Logistic Regression
(LogisticRegression), Decision Tree Forest (Ran-
domForestClassifier), Naive Bayes (GaussianNaive-
BayesClassifier), decision trees (DecisionTreeClas-
sifier) and Multi-layer Perceptron (MLPCLassifier).
When testing the algorithms, the proportion of train-
ing and testing sets was considered to be 80% training
and 20% testing, regardless of the data set used. The
obtained results are available in the Table 3, which
contains the comparative analysis of the metrics ob-
tained by us in relation to the analyzed works, and
Figure 1 contains the results obtained for the final data
set (Symcat38).
For the first data set addressed (Kaggle (Patil,
2024)), the accuracy of the algorithms used is 1.0,
which suggests overfitting (overestimation). We
compared the result thus obtained with the work
(Shedthi B et al., 2023), in which the authors observe
the same problem with overfitting, adapting the data
set through a procedure described briefly, in which the
symptoms ”that do not weigh majorly in the predic-
tion of diseases” are removed from dataset, also tak-
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
604

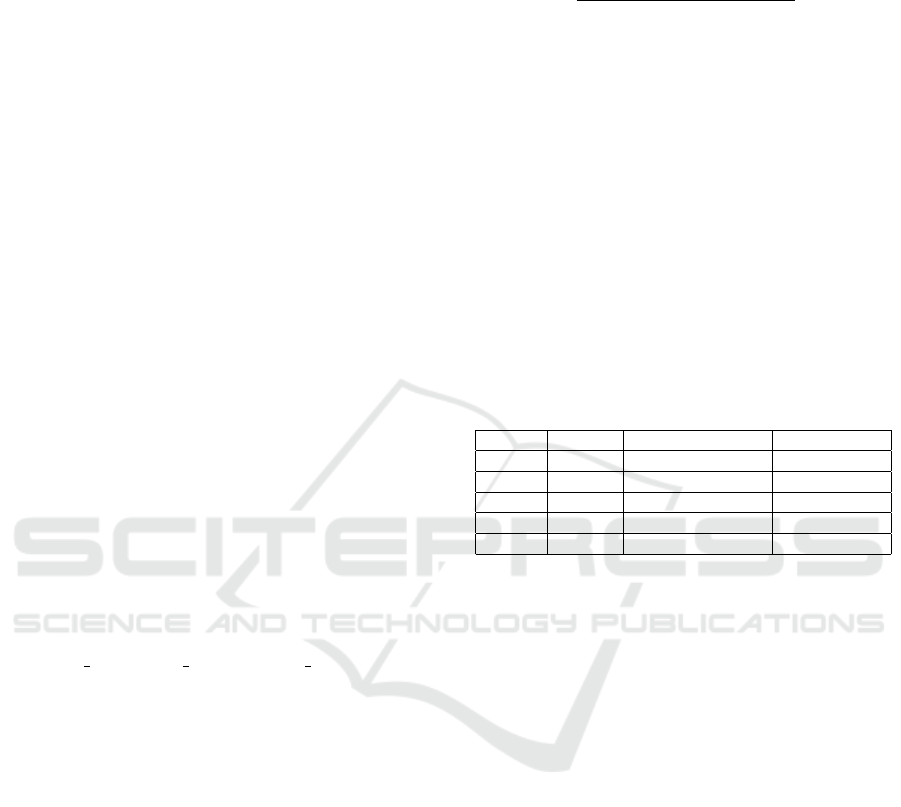
Table 3: Results obtained in the experiments.
Paper
DataSet
M1 M2 M3 M4 M5
Paper1
DataSet1
0.94 0.97 - - 0.96
Our Results
DataSet1
1.0 1.0 1.0 1.0 1.0
Paper2
DataSet2
- - - 0.55 -
Our Results
DataSet2
0.64 0.61 0.37 0.64 0.82
Paper3
DataSet3
0.6 0.58 - 0.60 0.60
Our Results
DataSet3
0.63 0.59 0.45 0.64 0.71
Our Results
DataSet4
0.72 0.71 0.53 0.72 0.80
ing into account the correlation between symptoms,
determined by a correlation matrix.
Algorithm testing on large datasets was done with
LargeDatasetHFAugumented (Anh, 2024), Medline-
Plus (Yuan and Yu, 2024) and Symcat (Yuan and Yu,
2024) (available at (Yuan and Yu, 2021)). The ac-
curacy of the algorithms is low. In the paper (Yuan
and Yu, 2024), the authors use their own disease pre-
diction algorithm based on Reinforcement Learning,
which aims to collect symptoms starting from an ini-
tial data set, finally performing the disease prediction
using MLPClassifier. The accuracy is 55% and they
are consistent with the similar results used in both the
paper (Polignano et al., 2020) and the experiment pro-
posed in this paper. Although other algorithms are
used in (Polignano et al., 2020), the accuracy does
not exceed 60%. The authors of the paper justify this
low level of accuracy on the grounds that in a data
set with many diseases and symptoms, the presented
models are not able to distinguish between diseases
that have a subset of common symptoms, but also the
fact that some diseases in the data set have vaguely de-
fined symptoms. A summative analysis of the exper-
iments is presented in Table 3 (with the mention that
”-” has been entered in the table for the untested algo-
rithms). In Table 3 the following notations were used:
DataSet1 is Kaggle, DataSet2 is Medline, DataSet3
is Symcat, DataSet4 is LargeDatasetHFAugumented
and Paper1 is (Shedthi B et al., 2023), Paper2 is
(Yuan and Yu, 2024) and Paper3 is (Polignano et al.,
2020). The notations from M1 to M5 represent the
methods applied in the order of their presentation in
the first paragraph of this chapter.
To avoid the problem of low accuracy in the case
of datasets with a large number of symptoms and dis-
eases, we manually selected a dataset consisting of
38 diseases and 167 symptoms (Symcat38), gathered
from the data available in the Git repository of the
paper (Yuan and Yu, 2024), several data files gener-
ated in Symcat, similar to the data used in (Polignano
et al., 2020). The selection of samples (patients) per
disease was carried out using a procedure similar to
that described in (Polignano et al., 2020), borrowed
from (Yuan and Yu, 2024). In this case, for a disease,
the prevalence of the disease is also known, i.e. for
each symptom in the list, a number between 0 and 1
is known, which represents the prevalence of the dis-
ease. To generate a synthetic patient, a list of values
following the uniform distribution is generated and a
boolean vector with the value 1 is formed if the gen-
erated value is lower than the prevalence value. Each
symptom present in the symptom list has an associated
real numerical value in the prevalence list. It is worth
mentioning that the prevalence value for a symptom is
different if it appears associated with several diseases.
To generate a synthetic patient for a disease, the pro-
cedure proceeds as follows: we assume that the list
of symptoms S has length l; This generates l real val-
ues between 0 and 1, by drawing l values from [0,
1) following the uniform distribution (ie each number
in the range has the same probability of being cho-
sen). Then, the prevalence value is subtracted from
each obtained value, and if the subtraction results in
a number less than 0 (the generated value for a symp-
tom is less than the prevalence value), then the symp-
tom will be added to the list of symptoms for the gen-
erated patient. The process is repeated until such a
list of length at least 2 is generated, since it would not
make sense to have diseases without associated symp-
toms in the data set, although it is possible in reality
in the case of asymptomatic patients, the situation is
difficult to model in the case classification algorithms
that require numerical data for training.
To generate patients for each disease, we gener-
ated 30 such patients, but after removing duplicates,
the number of patients per disease is varied. The re-
sulting dataset has 921 patients generated in total.
The algorithms used are the same, and the results
obtained are:
• Decision Trees - 0.61
• Random Forest - 0.720
• MultiLayer Perceptron - 0.78
• Gaussian Naive Bayes - 0.800
• Logistic regression - 0.805
Since the Logistic Regression algorithm has the
highest accuracy value, it is and the model we inte-
grated into the final application.
An important aspect to mention in the case of this
data set is the presence of two additional characteris-
Medical Chatbot for Disease Prediction Using Machine Learning and Symptom Analysis
605

Figure 1: The accuracy obtained for each algorithm.
tics compared to the others: age and sex. Experimen-
tally, we tried the problem of disease prediction on the
same data set, taking into account the generation of
age and sex. To encode the categories, in addition to
the one-hot vector obtained for the symptoms, the two
numerical values corresponding to the age and gender
categories are added. The performance of the algo-
rithms is comparable, in the case of GaussianNaive-
Bayes the accuracy even reaches 83%. However,
the obtained models are not capable of capturing all
the relationships between symptoms and sex/age, this
fact being highlighted in an example where the dis-
ease Uterine fibroids is predicted with a high thresh-
old (over 65%) in the case of all algorithms, even if
the selected gender is in category 0 (male) and given
that there is no training data in the dataset except for
symptoms and female gender. The problem can be ex-
plained by the representation of the data (symptoms,
age and sex), which is sparse in the case of symptoms,
and the other two values added to the obtained list do
not have such a significant weight in the model train-
ing phase.
For the construction of the initial model of the
chatbot, we proposed a model that starts from a set
of sentences/phrases, classifies them (using multi-
class classification) into categories of intentions (in-
tents), identifies entities with names (symptoms and
diseases), after which generates answers using the al-
gorithm for classifying symptoms into diseases and
additional information from a manually compiled
knowledge base.
Intent classification is a text classification problem
where the user’s phrases need to be classified into cer-
tain categories in order for the chatbot to provide an-
swers based on the task/question it needs to answer.
We manually generated a dataset where each intent
(tag) contains a list of sentences in json format. The
task of text classification starts with text preprocess-
ing, at which stage we used the NLTK (Natural Lan-
guage Toolkit) library (Bird et al., 2009) to remove
punctuation from texts, tokenize sentences and bring
words to a basic (dictionary-derived – “ stem”) us-
ing SnowBallStemmer. For the actual classification
model we used the Tensorflow platform (Abadi et al.,
2015). In the first phase, we used the Tokenizer class
to build a vocabulary and transform the processed
sentences into numerical form, and for the neural net-
work we chose a sequential architecture consisting of
layers of embedding, bidirectional lstm, dense (with
relu-enabled function), dropout and dense on the final
start (with the softmax activation function to obtain
the class/intent probabilities). Model training used
the crossentropy categorical loss function and Adam
as optimizer and a number of 100 epochs. The model
achieves 1.0 accuracy on the training and 0.77 on the
test data set.
6 CONCLUSIONS
The medical field is one of the most important fields
in everyday life, in this study we tried to demon-
strate that this vital field can be improved using ma-
chine learning together with other artificial intelli-
gence techniques. In the application developed in this
work, we demonstrated the construction of a medi-
cal chatbot capable of understanding the user’s intent,
extracting entities from texts and providing answers
based on information available in the form of csv files.
The chatbot made in this way represents a promising
application in the medical field, but which can be sig-
nificantly improved. First, the algorithms used to pre-
dict diseases based on symptoms could be replaced
by more sophisticated algorithms that ensure high ac-
curacy even when applied to larger datasets. Second,
the performance of such a model can be improved by
using qualitative data sets that contain more character-
istics besides symptoms and age/sex (gender), such as
symptom duration and risk factors. Furthermore, gen-
erative artificial intelligence models specialized in the
medical field could be used to generate the answers so
that the answers obtained are both scientifically cor-
rect and diverse to engage the user in conversation.
This paper underlines the significance of incorpo-
rating technology breakthroughs into medical prac-
tices by demonstrating the potential of machine learn-
ing approaches. It is essential to acknowledge this
study’s limitations. The quality and representative-
ness of the given datasets determine how accurate and
generalizable the classification models are.
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
606

Within this paper it was obtained satisfactory re-
sults, making a comparison with related work it can
be seen that the results obtained are good. The find-
ings of this research contribute to the growing body
of knowledge about machine learning applications in
the medical field and provide a base for future studies
aimed at improving medical practices and improving
communication with patients.
Future work would consist of creating a bigger
data set and testing and validating the models cre-
ated in this paper on this new data set, respectively,
trying to check what performance could be obtained
with other ML approaches.
REFERENCES
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z.,
Citro, C., Corrado, G. S., Davis, A., Dean, J., Devin,
M., Ghemawat, S., Goodfellow, I., Harp, A., Irving,
G., Isard, M., Jia, Y., Jozefowicz, R., Kaiser, L., Kud-
lur, M., Levenberg, J., Man
´
e, D., Monga, R., Moore,
S., Murray, D., Olah, C., Schuster, M., Shlens, J.,
Steiner, B., Sutskever, I., Talwar, K., Tucker, P., Van-
houcke, V., Vasudevan, V., Vi
´
egas, F., Vinyals, O.,
Warden, P., Wattenberg, M., Wicke, M., Yu, Y., and
Zheng, X. (2015). TensorFlow: Large-scale machine
learning on heterogeneous systems. Software avail-
able from tensorflow.org.
Almeida, L. B. (2020). Multilayer perceptrons. In Hand-
book of Neural Computation, pages C1–2. CRC Press.
Altamimi, I., Altamimi, A., Alhumimidi, A. S., Altamimi,
A., and Temsah, M.-H. (2023). Artificial intelligence
(ai) chatbots in medicine: a supplement, not a substi-
tute. Cureus, 15(6).
Anh, B. H. Q. (2024). Disease symptoms, @ONLINE.
Babu, A. and Boddu, S. B. (2024). Bert-based medical chat-
bot: Enhancing healthcare communication through
natural language understanding. Exploratory Re-
search in Clinical and Social Pharmacy, 13:100419.
Bali, M., Mohanty, S., Chatterjee, S., Sarma, M., and Pura-
vankara, R. (2019). Diabot: a predictive medical chat-
bot using ensemble learning. International Journal of
Recent Technology and Engineering, 8(2):6334–6340.
Bird, S., Klein, E., and Loper, E. (2009). Natural language
processing with Python: analyzing text with the natu-
ral language toolkit. ” O’Reilly Media, Inc.”.
Chakraborty, S., Paul, H., Ghatak, S., Pandey, S. K., Ku-
mar, A., Singh, K. U., and Shah, M. A. (2022). An
ai-based medical chatbot model for infectious disease
prediction. Ieee Access, 10:128469–128483.
Costa, V. G. and Pedreira, C. E. (2023). Recent advances
in decision trees: An updated survey. Artificial Intel-
ligence Review, 56(5):4765–4800.
Hapke, H., Howard, C., and Lane, H. (2019). Natural
Language Processing in Action: Understanding, an-
alyzing, and generating text with Python. Simon and
Schuster.
Karuna, G., Reddy, G. G., Sushmitha, J., Gayathri, B.,
Sharma, S. D., and Khatua, D. (2023). Helpi–an auto-
mated healthcare chatbot. In E3S Web of Conferences,
volume 430, page 01040. EDP Sciences.
Patil, P. (2024). Symptoms and diseases dataset @ON-
LINE.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer,
P., Weiss, R., Dubourg, V., et al. (2011). Scikit-
learn: Machine learning in python. Journal of ma-
chine learning research, 12(Oct):2825–2830.
Polignano, M., Narducci, F., Iovine, A., Musto, C.,
De Gemmis, M., and Semeraro, G. (2020). Healthas-
sistantbot: a personal health assistant for the italian
language. IEEE Access, 8:107479–107497.
Probst, P., Wright, M. N., and Boulesteix, A.-L. (2019). Hy-
perparameters and tuning strategies for random for-
est. Wiley Interdisciplinary Reviews: data mining and
knowledge discovery, 9(3):e1301.
Reddy, E. M. K., Gurrala, A., Hasitha, V. B., and Kumar, K.
V. R. (2022). Introduction to naive bayes and a review
on its subtypes with applications. Bayesian reasoning
and gaussian processes for machine learning applica-
tions, pages 1–14.
Shedthi B, S., Shetty, V., Chadaga, R., Bhat, R., Bangera, P.,
and Kini K, P. (2023). Implementation of chatbot that
predicts an illness dynamically using machine learn-
ing techniques. International Journal of Engineering,
(Articles in Press).
Siddique, S. and Chow, J. C. (2021). Machine learning in
healthcare communication. Encyclopedia, 1(1):220–
239.
Tode, V., Gadge, H., Madane, S., Kachare, P., and Deokar,
A. (2021). A chatbot for medical purpose using deep
learning. International Journal of Engineering Re-
search & Technology.
Vasileiou, M. V. and Maglogiannis, I. G. (2022). The health
chatbots in telemedicine: Intelligent dialog system for
remote support. Journal of Healthcare Engineering,
2022(1):4876512.
Yuan, H. and Yu, S. (2021). Efficient symptom inquiring
and diagnosis via adaptive alignment of reinforcement
learning and classification.
Yuan, H. and Yu, S. (2024). Efficient symptom inquiring
and diagnosis via adaptive alignment of reinforcement
learning and classification. Artificial Intelligence in
Medicine, 148:102748.
Zabor, E. C., Reddy, C. A., Tendulkar, R. D., and Patil, S.
(2022). Logistic regression in clinical studies. In-
ternational Journal of Radiation Oncology* Biology*
Physics, 112(2):271–277.
Zhang, Y., Chen, Q., Yang, Z., Lin, H., and Lu, Z. (2019).
Biowordvec, improving biomedical word embeddings
with subword information and mesh. Scientific data,
6(1):52.
Medical Chatbot for Disease Prediction Using Machine Learning and Symptom Analysis
607
