
A Multifractal-Based Masked Auto-Encoder: An Application to Medical
Images
Joao Batista Florindo
a
and Viviane de Moura
Institute of Mathematics, Statistics and Scientific Computing, University of Campinas, Rua S
´
ergio Buarque de Holanda,
651, Cidade Universit
´
aria ”Zeferino Vaz” - Distr. Bar
˜
ao Geraldo, CEP 13083-859, Campinas, SP, Brazil
fl
Keywords:
Masked Auto-Encoder, Multifractal Spectrum, Medical Images.
Abstract:
Masked autoencoders (MAE) have shown great promise in medical image classification. However, the ran-
dom masking strategy employed by traditional MAEs may overlook critical areas in medical images, where
even subtle changes can indicate disease. To address this limitation, we propose a novel approach that utilizes
a multifractal measure (Renyi entropy) to optimize the masking strategy. Our method, termed Multifractal-
Optimized Masked Autoencoder (MO-MAE), employs a multifractal analysis to identify regions of high com-
plexity and information content. By focusing the masking process on these areas, MO-MAE ensures that the
model learns to reconstruct the most diagnostically relevant features. This approach is particularly beneficial
for medical imaging, where fine-grained inspection of tissue structures is crucial for accurate diagnosis. We
evaluate MO-MAE on several medical datasets covering various diseases, including MedMNIST and COVID-
CT. Our results demonstrate that MO-MAE achieves promising performance, surpassing other basiline and
state-of-the-art models. The proposed method also adds minimum computational overhead as the computa-
tion of the proposed measure is straightforward. Our findings suggest that the multifractal-optimized masking
strategy enhances the model’s ability to capture and reconstruct complex tissue structures, leading to more
accurate and efficient medical image representation. The proposed MO-MAE framework offers a promising
direction for improving the accuracy and efficiency of deep learning models in medical image analysis, poten-
tially advancing the field of computer-aided diagnosis.
1 INTRODUCTION
Self-supervised learning (SSL) has emerged as a pow-
erful paradigm in modern deep learning, offering a
promising approach to overcome the limitations of
traditional supervised and unsupervised methods (Do-
ersch and Zisserman, 2017). The approach has gained
significant traction in recent years, particularly in do-
mains such as computer vision and applications as in
computer-aided diagnostics (Krishnan et al., 2022).
Masked Autoencoder (MAE) (He et al., 2022) is an
example of well-succeeded SSL method. MAEs work
by reconstructing images from partially masked in-
puts, encouraging the model to learn meaningful rep-
resentations by aggregating contextual information.
However, the random masking strategy employed
by traditional MAEs may not be optimal for medical
images, where subtle changes in specific regions can
be crucial for accurate diagnosis. In medical imaging,
certain areas often contain more diagnostically rele-
a
https://orcid.org/0000-0002-0071-0227
vant information than others. For instance, in chest
X-rays, the lung fields typically hold more critical in-
formation for detecting respiratory diseases compared
to the surrounding areas. To address this limitation,
researchers have explored approaches to optimize the
masking strategy of MAEs, also in applications to
medical images (Mao et al., 2024).
One promising direction to quantify the relevance
of image regions and consequently guide MAE mask-
ing process, and that has not yet been explored in
the literature for this purpose, is multifractal analysis.
This has been successfully applied in image process-
ing and pattern recognition applications, e.g., in tex-
ture analysis and classification (Florindo and Neckel,
2023). Multifractal analysis provides a framework
for describing the complexity and heterogeneity of
images across different scales, making it particularly
suitable for capturing the intricate structures often
present in real-world images. One of the most ef-
fective and straightforward techniques for multifrac-
tal analysis in digital images is Renyi entropy (R
´
enyi,
Florindo, J. B. and de Moura, V.
A Multifractal-Based Masked Auto-Encoder: An Application to Medical Images.
DOI: 10.5220/0013359300003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 3: VISAPP, pages
769-776
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
769

1961). This is a generalization of Shannon entropy
and has been used in image processing, for exam-
ple in texture recognition (Florindo, 2023). Its abil-
ity to characterize the information content of images
at different scales makes it a potential candidate for
optimizing masking strategies in MAEs for medical
imaging.
Building upon these foundations, this paper intro-
duces a novel approach that combines the strengths
of MAEs with multifractal analysis to enhance med-
ical image classification. By utilizing Renyi entropy
as a multifractal measure to guide the masking pro-
cess, our proposed Multifractal-Optimized Masked
Autoencoder (MO-MAE) aims to focus on regions
of high complexity and information content, ensuring
that the model learns to reconstruct the most diagnos-
tically relevant features. Our main contributions and
innovations are as follows:
• We develop a multifractal-based masking strat-
egy for MAE, improving results on medical im-
age classification in the literature; our approach
can also be easily extended to other application
domains, in general tasks related to image classi-
fication.
• Up to our knowledge, this is the first time that
multifractal analysis (and Renyi entropy in partic-
ular) is associated with masked auto-encoders in
the literature.
• Being even more general, this is the first time that
a physics-based complexity measure is explored
in the MAE masking process, as other masking
strategies usually rely on learnable procedures.
• We assess the proposed methodology on the
well-established benchmarks of medical images
MedMNIST (Yang et al., 2023) as well as on
the real-world task of predicting COVID cases -
dataset COVID-CT (Yang et al., 2020). Exten-
sive evaluations and comparison with results re-
cently published in the literature are performed
over those databases to confirm the potential of
our proposal.
The proposed MO-MAE model outperforms other
literature approaches in most scenarios both in the
benchmark datasets and on the COVID-CT problem.
Overall, our results suggest that using multifractal
analysis to guide the masking strategy in the MAE
framework is a promising direction and can be fur-
ther explored, including applications to other domains
outside the medical field or even other tasks, such as
segmentation, for instance.
2 RELATED WORKS
Masked Autoencoders (MAE) have emerged as a
promising paradigm for self-supervised learning in
computer vision, achieving state-of-the-art perfor-
mance across various benchmark datasets (He et al.,
2022). MAEs have also been investigated in med-
ical applications, particularly in image analysis and
classification tasks. For example, in electrocardio-
graphy analysis, MAE-based self-supervised learn-
ing has shown promise in improving model perfor-
mance for detecting left ventricular systolic dysfunc-
tion, even with limited training data (Sawano et al.,
2024). An overview on this topic can be found in (Kr-
ishnan et al., 2022).
Improvements over the original MAE architecture
have also been explored. Several of them have fo-
cused on more elaborate masking strategies. That is
the case of (Bandara et al., 2023), where an adaptive
masking is proposed, using an auxiliary network that
samples visible tokens based on the semantic context.
Another one is (Li et al., 2022), where the authors
propose a semantic-guided masking strategy. This is
achieved by encouraging the neural network to learn
various information from intra-part patterns to inter-
part relations. A study specifically focused on medi-
cal images is described in (Mao et al., 2024), where
the authors propose the use of attention maps obtained
by a supervised procedure to conduct the masking
process. Theoretical studies on the role of masking in
MAEs have also been presented, as in (Zhang et al.,
2022). Our proposal goes in another direction here
as we focus on the use of a complexity measure to
guide the masking process. Among the advantages of
such approach, we can mention the interpretability of
the masking criterion and the fact that our model does
not require the learning of extra parameters in the pre-
training stage or any other external training algorithm.
Fractal and multifractal theory have also been ex-
plored in the literature of image analysis, especially
in medical images. In (Ding et al., 2023), a frac-
tal graph convolutional neural network is proposed
for computer-aided diagnosis using histopathological
images. In (Swapnarekha et al., 2024), a review of
fractal-based image analysis with pattern recognition
is presented. The authors in (Motwani and Fadnavis,
2024) investigate the correlation between the fractal
dimension computed on CBCT scans of edentulous
patients on implant site with the bone density deter-
mined by Misch’s classification. Renyi entropy, par-
ticularly, was explored for image classification, for
example, in (Florindo, 2023), where it was employed
to analyze representations of deep convolutional neu-
ral networks. A combination of multifractal analysis
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
770

with stacked autoencoders (not masked) is reported
in (Yu et al., 2022), where multifractal theory is used
for feature learning. The use of multifractal analysis
to guide MAE masking strategy is a novelty of our
study.
3 PROPOSED METHOD
3.1 Overall Model
Despite the effectiveness of masked autoencoders in
the literature, space for improvements still exist. One
of such possibilities concern the mask selection step.
Although the well-established random masking ap-
proach is straightforward, it does not take into account
particularities of each image, for example, regions
with more or less relevant information. In this con-
text, here we present MO-MAE, a novel approach to
MAE masking using multifractal analysis. Multifrac-
tal theory was originally developed to analyze com-
plex physical systems, but has also demonstrated po-
tential in image analysis (Florindo and Neckel, 2023),
particularly quantifying the complexity of image re-
gions and, as a consequence, its importance for the
global image representation.
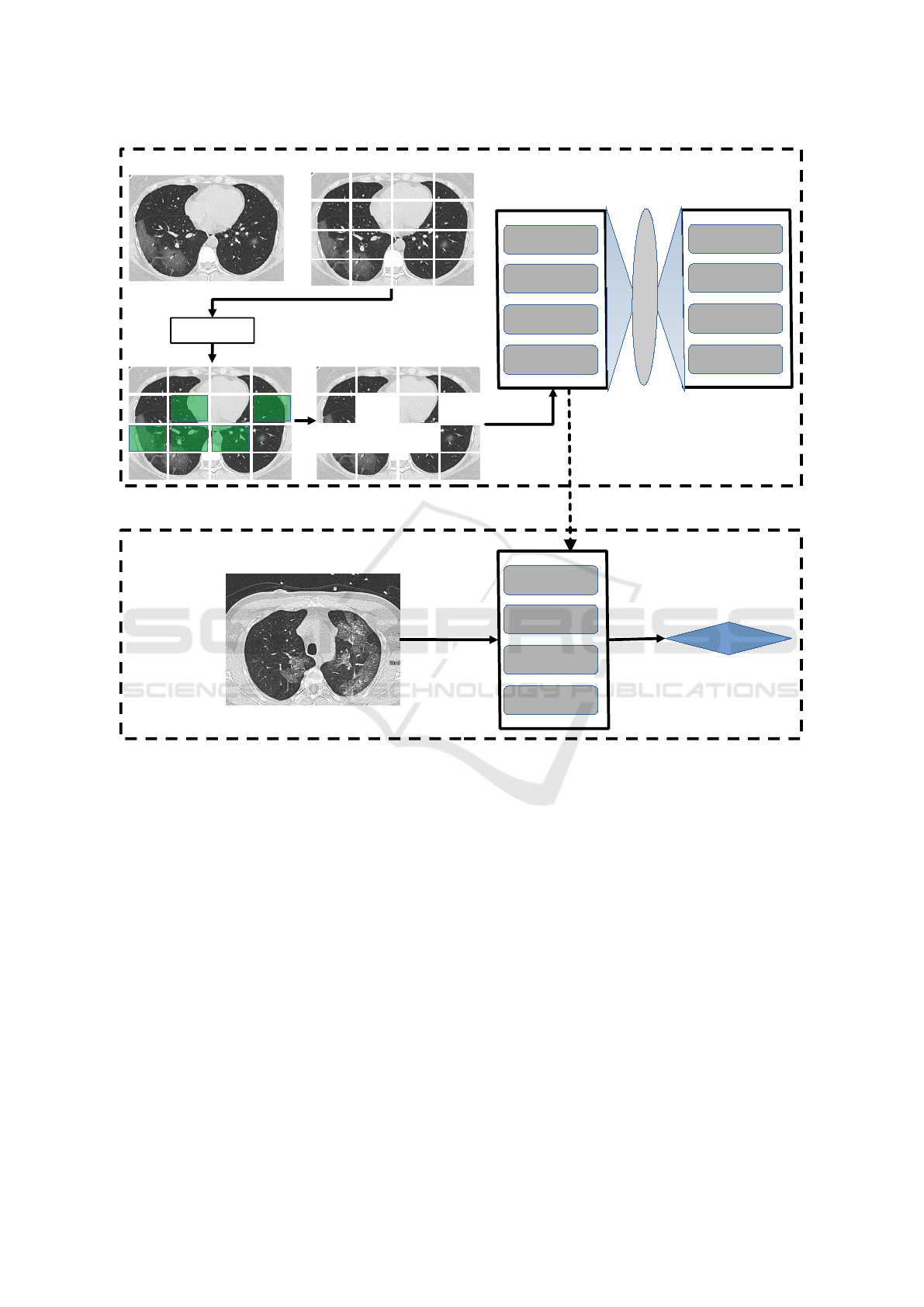
Figure 1 provides a general schematic overview
of the proposed methodology. As usual in self-
supervised frameworks, the architecture is divided
at high level into two tasks: the pretraining (up-
stream task) and fine-tuning (downstream). The over-
all model comprises the following major sequential
steps:
1. Patching: The image is partitioned into a collec-
tion of rectangular patches. The number and size
of patches are hyperparameters to be determined
by the user.
2. Multifractal Analysis: The multifractal spec-
trum is computed over each patch (more details
on that in Section 3.2).
3. Masking: Based on the multifractal spectrum, we
select those patches with large amount of useful
information. The percentage of selected patches
is a hyperparameter.
4. Encoder/Decoder Pretraining: The selected
patches are used as input to an encoder module,
which is a Vision Transformer (ViT). This is re-
sponsible for providing a latent representation of
the input with reduced dimensionality. The output
of the encoder is the input of another ViT, which
plays the role of decoder. Both encoder and de-
coder are jointly trained with the objective of re-
constructing the original image from the patches
selected by the multifractal spectrum. The loss
function measures the discrepancy between the
original and the reconstructed images, as in (He
et al., 2022).
5. Prediction (Fine-Tuning): Finally, the model re-
ceives the images of the target task (e.g., the diag-
nostic of a disease), previously labeled by a spe-
cialist or any other exogenous mechanism (e.g.,
a genetic test). This is processed by the encoder
pretrained on the reconstruction task and this en-
coder is fine-tuned over the new labeled images.
The final model is ready to be used on the test set
and in the real-world application.
3.2 Multifractal Analysis
Our main novelty lies in the pretraining stage, in par-
ticular, in the multifractal selector, responsible for
defining the patches that will be used as input to
the reconstruction task. In (Falconer, 2013), two
types of spectra are defined for multifractal analy-
sis: the singularity and coarse spectra. For image
analysis, given the limitation of the multiscale anal-
ysis imposed by the underlying resolution, the first
one is more usual in general. Theoretical details can
be found, for instance, in (Falconer, 2013), but in
computational terms, we employ the partition func-
tion method (Salat et al., 2017). Assuming a single-
channel image I : Z
2
→ Z, the codomain is partitioned
with even spacing s, giving rise to
y
j
(s) =
js
∑
i=( j−1)s+1
I(I(·) = i), 1 ≤ j ≤ N
s
= ⌊G/s⌋,
where I is the indicator function and G is the number
of pixel intensity levels (default 255). From that, we
estimate the probability distribution
p
j
(s) =
y
j
(s)
∑
N
s
k=1
y
k
(s)
.
The q-th moment partition function is therefore de-
fined by
Z
q
s
=
N
s
∑
j=1
[p
j
(s)]
q
,
which in a multifractal regime should obey the fol-
lowing power-law rule:
Z
q
s
∼ s
τ(q)
.
τ(q) is the scaling exponent function, also known as
the multifractal spectrum of I. This also gives rise to
an associated entropy, which is Renyi entropy, defined
by
R
q
s
=
1
1 − q
log(Z
q
s
).
A Multifractal-Based Masked Auto-Encoder: An Application to Medical Images
771
Multifractal
Selector
TB4
TB3
TB2
TB1
ENCODER
TB4
TB3
TB2
TB1
DECODER
L
a
t
e
n
t
R
e
a
p
r
e
s
e
n
t
a
t
i
o
n
TB4
TB3
TB2
TB1
Transfer Learning
Prediction
PRETRAINING
FINE-TUNING
Figure 1: Proposed method. In the pretraining stage (upper block) we start by partitioning the image into rectangular patches
(the number of patches here is only for illustrative purposes). Therefore we apply the multifractal selector module to identify
patches with sufficiently relevant information. This is used as input to a ViT encoder, comprising 4 Transformer Blocks (TB).
A mirrored architecture is used for decoding. The pretrained encoder is used in the fine-tuning step (lower block), to provide
the deep latent representation of the input image and provide the desired prediction.
The case q = 1 is defined as being equivalent to the
well-known Shannon entropy.
Here we divide the input image I : Z
M×N
→ Z
into N
′
× N
′
non-overlapping patches. The number
of patches is n
P
= ⌊N/N
′
⌋ × ⌊M/N
′
⌋. For the k
th
patch P
k
we compute the Renyi entropy R
q
s
(P
k
). The
patches are sorted in descendant order according to
the entropy. Formally, let P = (P
k
)
n
P
k=1
be a sequence
of patches such that R
q
s
(P
k+1
) ≥ R
q
s
(P
k
). Provided the
mask ratio r ∈ [0, 1], the number of selected patches is
n
S
= (1 − r)n
P
and the patches are obtained from the
subsequence
P
′
=
P
n
k
n
k
={1,2,3,···,n
S
}
.
The set of selected patches P
′
is finally introduced
as input to the encoder in the pre-training state and all
the remaining steps follows as described in Section
3.1. We ensure in this way that those patches with
high complexity, as measured by Renyi entropy, are
selected for the reconstruction. These also correspond
to the richest regions on the image, and consequently
those parts whose reconstruction is more challenging.
By forcing the pretraining encoder to solve such dif-
ficult task, we gather more robust and richer features
in the latent representation, which naturally will lead
to more effectiveness in the target task, image classi-
fication in our case.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
772

4 EXPERIMENTAL SETUP
For the implementation of the MAE algorithm, we
adopted a patch size of 16 × 16, 4 layers both in the
encoder and decoder ViT, 200 epochs in the pretrain-
ing stage and 100 epochs for fine-tuning. The num-
ber of layers and pretraining epochs are considerably
smaller than the original model, which used 12 lay-
ers and 2000 epochs, respectively. We observed that
enlarging the backbone did not correspond to any sig-
nificant improvement for our purposes. For the re-
maining hyperparameters we adopted default values,
using AdamW as the optimizer. In the pretraining,
we used a base learning rate of 1.5e-4, weight decay
0.05, batch size 4096 (MedMNIST) or 128 (COVID-
CT), and mask ratio 0.75 (75% of patches masked).
In the fine-tuning, we used a base learning rate of 1e-
3, weight decay 0.5, and batch size 128. The experi-
ments were carried out on Google Colab environment
with an Nvidia T4 GPU.
The performance of the proposed methodology
was assessed on the collection of medical image
datasets MedMNIST-V2 (Yang et al., 2023) and the
COVID-CT database (Yang et al., 2020). MedMNIST
is a family of datasets, including both 2D and 3D
biomedical images especially designed and prepro-
cessed for benchmark. Here we use the 2D collection,
which comprises a total of 708,000 labeled images,
each one with size 224 × 224. Those images cover
a wide range of medical modalities (pathology, X-
ray, dermatoscopy, retinal OCT, abdominal CT, breast
ultrasound, etc.) and predictive tasks (binary/multi-
class, ordinal regression, and multi-label). COVID-
CT, on the other hand, consists of 349 COVID-19 CT
and 397 Non-COVID-19 CT images. Those images
were resized to 224 × 224. The dataset was split into
a training, a validation, and a test set, by patient, with
a ratio of 60%, 15%, and 25%, respectively.
As comparative metrics, we adopt accuracy
(ACC), which is defined as the ratio of images cor-
rectly classified, area under the precision/recall curve
(AUC), and F1 score (harmonic mean of precision and
recall).
5 RESULTS AND DISCUSSION
5.1 MedMNIST
Table 1 presents the results of an ablation study, where
we compare the model with and without the multi-
fractal MAE module on MedMNIST datasets. We ob-
serve a general increase both in terms of accuracy and
AUC. This is even more evident in the most challeng-
ing data, as in RetinaMNIST and BreastMNIST, but
the superiority is consistent across all datasets.
Another important investigation concerns the role
of q hyperparameter in the multifractal spectrum
patch selection. This experiment is summarized by
Table 2. The values of q ∈ {1,2, 10} were chosen
from the empirical observation that other intermedi-
ate or larger/smaller values did not provide significant
difference in the final results. A classical intuition in
multifractal theory relates q with the role of a “magni-
fying glass”: larger values of q correspond to coarser
scales of analysis. Here we see that q = 2 is in general
a compromise between short and long-range fractality
observed over the patch. Based on that, this was our
choice for the remaining experiments.
Table 3 lists results recently published in the lit-
erature on MedMNIST datasets in comparison with
the proposed approach. MO-MAE attains the high-
est AUC/ACC in most datasets. Here AUC is a
more faithful metric considering possible imbalances
in some of those datasets. And, with respect to AUC,
the only exceptions where MO-MAE is the not the
best method are PneumoniaMNIST, BreastMNIST,
TissueMNIST, and OrganSMNIST. In all these cases,
the highest AUC corresponds to MedVIT-S (Manzari
et al., 2023). We should highlight, however, that this
is a computationally intensive architecture from the
state-of-the-art, combining deep Convolutional Neu-
ral Networks and Transformers. And even in these
scenarios, our results are quite competitive. And it
is also interesting to observe that MO-MAE outper-
formed MedVIT in most datasets, even considering
the largest version MedVIT-L. Another point that is
worth it to mention is the lack of competitiveness of
fully automatic methods, such as Auto-sklearn, Au-
toKeras, and Google AutoML. Our results confirm
that deep learning algorithms appropriately tuned for
each particular task still is the best option in most
practical situations.
5.2 COVID-CT
Figure 2 depicts the precision/recall curve for the pro-
posed method on the COVID-CT database. Table 4
presents a comparison of our results on the COVID
data with the literature. The curve is in line with the
reported F1 score of 0.85 and follows a characteristic
pattern where low recall also corresponds to low pre-
cision. This behavior is typically observed in nearly-
balanced databases, which is the case of COVID-CT.
Figure 3 depicts the confusion matrix for MO-
MAE on the COVID-CT dataset. We notice that our
method performs well both with respect to the posi-
A Multifractal-Based Masked Auto-Encoder: An Application to Medical Images
773
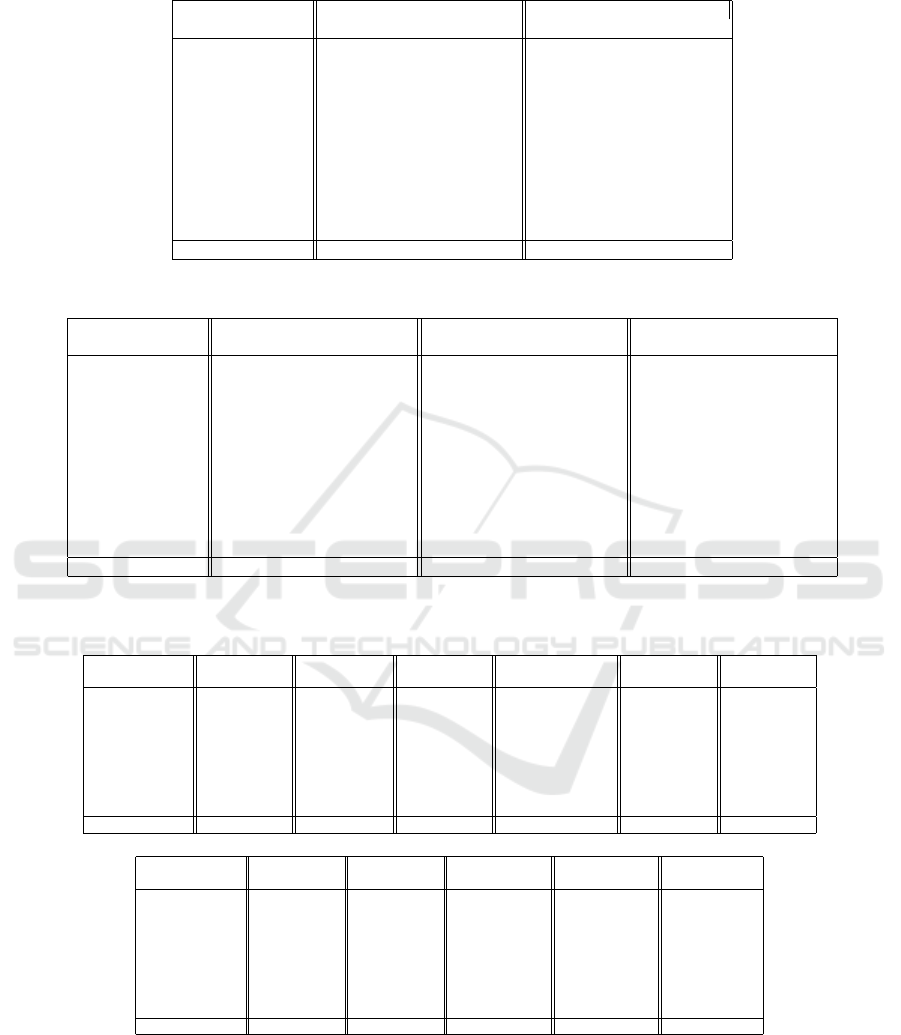
Table 1: Ablation experiment on MedMNIST datasets. The original MAE with classical masking strategy is compared with
the multifractal-guided approach proposed here.
Dataset Original MO-MAE (Proposed)
AUC ACC AUC ACC
PathMNIST 0.996 0.948 0.997 0.953
DermaMNIST 0.922 0.806 0.959 0.810
OCTMNIST 0.992 0.917 0.993 0.917
PneumoniaMNIST 0.936 0.910 0.988 0.909
RetinaMNIST 0.734 0.497 0.822 0.588
BreastMNIST 0.855 0.885 0.920 0.872
BloodMNIST 0.999 0.989 1.000 0.988
TissueMNIST 0.930 0.709 0.944 0.720
OrganAMNIST 0.998 0.966 0.999 0.959
OrganCMNIST 0.995 0.941 0.998 0.939
OrganSMNIST 0.982 0.834 0.983 0.831
Average 0.940±0.082 0.855±0.144 0.964±0.054 0.862±0.120
Table 2: Evaluation of hyperparameter q on MedMNIST datasets.
Dataset q = 1 q = 2 q = 10
AUC ACC AUC ACC AUC ACC
PathMNIST 0.994 0.930 0.997 0.953 0.996 0.948
DermaMNIST 0.931 0.762 0.959 0.810 0.929 0.799
OCTMNIST 0.971 0.789 0.993 0.917 0.992 0.917
PneumoniaMNIST 0.978 0.929 0.988 0.909 0.955 0.909
RetinaMNIST 0.715 0.502 0.822 0.588 0.731 0.492
BreastMNIST 0.898 0.840 0.920 0.872 0.857 0.891
BloodMNIST 0.998 0.963 1.000 0.988 0.999 0.989
TissueMNIST 0.935 0.695 0.944 0.720 0.930 0.710
OrganAMNIST 0.998 0.948 0.999 0.959 0.999 0.967
OrganCMNIST 0.996 0.931 0.998 0.939 0.997 0.942
OrganSMNIST 0.982 0.821 0.983 0.831 0.983 0.836
Average 0.949±0.081 0.828±0.139 0.964±0.054 0.862±0.120 0.942±0.083 0.854±0.145
Table 3: Results for the proposed MO-MAE method on MedMNIST datasets compared with other methods in the literature.
Literature results obtained from (Manzari et al., 2023).
Method PathMNIST DermaMNIST OCTMNIST PneumoniaMNIST RetinaMNIST BreastMNIST
AUC ACC AUC ACC AUC ACC AUC ACC AUC ACC AUC ACC
ResNet-18 0.989 0.909 0.920 0.754 0.958 0.763 0.956 0.864 0.710 0.493 0.891 0.833
ResNet-50 0.989 0.892 0.912 0.731 0.958 0.776 0.962 0.884 0.716 0.511 0.866 0.842
Auto-sklearn 0.934 0.716 0.902 0.719 0.887 0.601 0.942 0.855 0.690 0.515 0.836 0.803
AutoKeras 0.959 0.834 0.915 0.749 0.955 0.763 0.947 0.878 0.719 0.503 0.871 0.831
Google AutoML 0.944 0.728 0.914 0.768 0.963 0.771 0.991 0.946 0.750 0.531 0.919 0.861
MedVIT-T 0.994 0.938 0.914 0.768 0.961 0.767 0.993 0.949 0.752 0.534 0.934 0.896
MedVIT-S 0.993 0.942 0.937 0.780 0.960 0.782 0.995 0.961 0.773 0.561 0.938 0.897
MedVIT-L 0.984 0.984 0.920 0.773 0.945 0.761 0.991 0.921 0.754 0.552 0.929 0.883
MO-MAE 0.997 0.953 0.959 0.810 0.993 0.917 0.988 0.909 0.822 0.588 0.920 0.872
Method BloodMNIST TissueMNIST OrganAMNIST OrganCMNIST OrganSMNIST
AUC ACC AUC ACC AUC ACC AUC ACC AUC ACC
ResNet-18 0.998 0.963 0.933 0.681 0.998 0.951 0.994 0.920 0.974 0.778
ResNet-50 0.997 0.950 0.932 0.680 0.998 0.947 0.993 0.911 0.975 0.785
Auto-sklearn 0.984 0.878 0.828 0.532 0.963 0.762 0.976 0.829 0.945 0.672
AutoKeras 0.998 0.961 0.941 0.703 0.994 0.905 0.990 0.879 0.974 0.813
Google AutoML 0.998 0.966 0.924 0.673 0.990 0.886 0.988 0.877 0.964 0.749
MedVIT-T 0.996 0.950 0.943 0.703 0.995 0.931 0.991 0.901 0.972 0.789
MedVIT-S 0.997 0.951 0.952 0.731 0.996 0.928 0.993 0.916 0.987 0.805
MedVIT-L 0.996 0.954 0.935 0.699 0.997 0.943 0.994 0.922 0.973 0.806
MO-MAE 1.000 0.988 0.944 0.720 0.999 0.959 0.998 0.939 0.983 0.831
tive and negative classes.
Table 4 lists some results published in the litera-
ture for the COVID-CT database in comparison with
MO-MAE in terms of accuracy and F1 score. Here
we follow the protocol in (Abid et al., 2023), which
does not involve any pre-segmentation task, and con-
sequently is more challenging that that explored in
the original reference (Yang et al., 2020). That is
the reason why our method is not comparable with
(Yang et al., 2020) but with (Abid et al., 2023). MO-
MAE achieves significant advantage, even over mod-
els with huge number of learnable parameters, such as
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
774
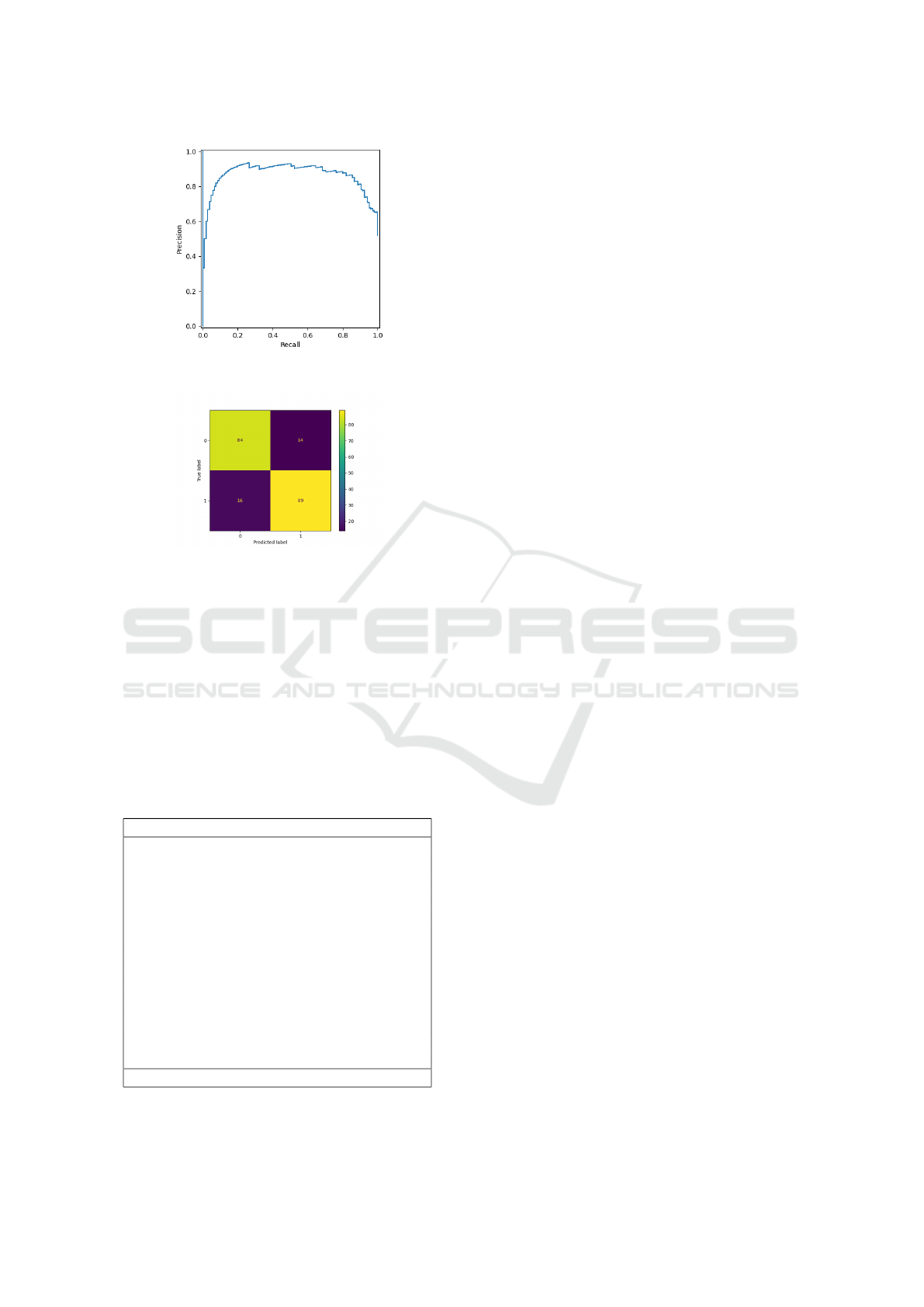
Figure 2: Precision/Recall curve for the proposed MO-
MAE method on the COVID-CT dataset.
Figure 3: Confusion matrix for the proposed MO-MAE
method on the COVID-CT dataset.
DenseNet-169 and ResGANet-101. Another notice-
able point is how most standard CNNs do not achieve
good performance even using transfer learning strate-
gies. COVID-CT images present particular subtleties
that can be hardly learned without the introduction of
extra prior information. Our self-supervised approach
demonstrates to be a promising solution in this direc-
tion.
Table 4: Results for MO-MAE on COVID-CT dataset com-
pared with other methods in the literature. Literature results
obtained from (Abid et al., 2023).
Method Accuracy F1 Score
VGG-16 0.66 0.58
ResNet-50 0.72 0.73
DenseNet-169 0.80 0.79
EfficientNet-b1 0.70 0.62
CRNet 0.72 0.76
ShuffleNetV2 (1.5X) 0.73 0.76
SENet-50 0.76 0.77
CBAM-50 0.78 0.80
ResNeXt-50 0.72 0.75
Res2Net-50 0.73 0.74
ECANet-50 0.75 0.74
SKNet-50 0.77 0.76
ResGANet-101 (G=2) 0.78 0.81
MO-MAE 0.85 0.85
Overall, the presented results suggest the poten-
tial of the proposed MO-MAE model as a power-
ful solution for medical image classification. The
method outperformed several models with high com-
putational burden in the literature and also demon-
strated stability and robustness across different types
of images and medical tasks.
6 CONCLUSIONS
In this work, we proposed a new strategy for masking
in masked auto-encoders. The masking process was
guided by the multifractal spectrum computed over
the image patches. Patches with the highest Renyi
entropies were selected to compose the input to the
pretraining task.
The efficiency of our proposal was assessed on
benchmarks of medical images commonly used in
the literature: MedMNIST collection of datasets and
COVID-CT database. The obtained results are en-
couraging, demonstrating competitiveness with the
state-of-the-art on medical image classification using
deep learning. Particularly, our approach follows the
self-supervised paradigm, which also makes it a natu-
rally interesting solution in scenarios where the num-
ber of labeled images for training is limited. This is
especially common in several areas of medicine.
Our approach can also be straightforwardly ex-
tended to other domains involving image classifica-
tion and even related tasks, such as segmentation,
for example. The proposed method might also ben-
efit from the use of larger datasets, given that this is
the scenario where self-supervised learning typically
stands out. Future investigation on these possibilities
are in progress.
ACKNOWLEDGEMENTS
Joao Batista Florindo gratefully acknowledges the fi-
nancial support of the S
˜
ao Paulo Research Foundation
(FAPESP) (Grants #2024/01245-1 and #2020/09838-
0) and from National Council for Scientific and
Technological Development, Brazil (CNPq) (Grant
#306981/2022-0).
REFERENCES
Abid, M. H., Ashraf, R., Mahmood, T., and Faisal, C. N.
(2023). Multi-modal medical image classification us-
ing deep residual network and genetic algorithm. Plos
one, 18(6):e0287786.
A Multifractal-Based Masked Auto-Encoder: An Application to Medical Images
775

Bandara, W. G. C., Patel, N., Gholami, A., Nikkhah, M.,
Agrawal, M., and Patel, V. M. (2023). Adamae:
Adaptive masking for efficient spatiotemporal learn-
ing with masked autoencoders. In Proceedings of the
IEEE/CVF Conference on Computer Vision and Pat-
tern Recognition, pages 14507–14517.
Ding, S., Gao, Z., Wang, J., Lu, M., and Shi, J. (2023).
Fractal graph convolutional network with mlp-mixer
based multi-path feature fusion for classification of
histopathological images. Expert Systems with Appli-
cations, 212:118793.
Doersch, C. and Zisserman, A. (2017). Multi-task self-
supervised visual learning. In Proceedings of the
IEEE international conference on computer vision,
pages 2051–2060.
Falconer, K. (2013). Fractal geometry: mathematical foun-
dations and applications. John Wiley & Sons.
Florindo, J. B. (2023). Renyi entropy analysis of a deep con-
volutional representation for texture recognition. Ap-
plied Soft Computing, 149:110974.
Florindo, J. B. and Neckel, A. (2023). A randomized net-
work approach to multifractal texture descriptors. In-
formation Sciences, 648:119544.
He, K., Chen, X., Xie, S., Li, Y., Doll
´
ar, P., and Girshick,
R. (2022). Masked autoencoders are scalable vision
learners. In Proceedings of the IEEE/CVF conference
on computer vision and pattern recognition, pages
16000–16009.
Krishnan, R., Rajpurkar, P., and Topol, E. J. (2022). Self-
supervised learning in medicine and healthcare. Na-
ture Biomedical Engineering, 6(12):1346–1352.
Li, G., Zheng, H., Liu, D., Wang, C., Su, B., and Zheng,
C. (2022). Semmae: Semantic-guided masking for
learning masked autoencoders. Advances in Neural
Information Processing Systems, 35:14290–14302.
Manzari, O. N., Ahmadabadi, H., Kashiani, H., Shokouhi,
S. B., and Ayatollahi, A. (2023). Medvit: a ro-
bust vision transformer for generalized medical image
classification. Computers in Biology and Medicine,
157:106791.
Mao, J., Guo, S., Yin, X., Chang, Y., Nie, B., and Wang,
Y. (2024). Medical supervised masked autoencoder:
Crafting a better masking strategy and efficient fine-
tuning schedule for medical image classification. Ap-
plied Soft Computing, page 112536.
Motwani, M. B. and Fadnavis, A. M. (2024). Fractal di-
mension analysis at implant site on cbct. International
Dental Journal, 74:S75.
R
´
enyi, A. (1961). On measures of entropy and informa-
tion. In Proceedings of the fourth Berkeley sympo-
sium on mathematical statistics and probability, vol-
ume 1: contributions to the theory of statistics, vol-
ume 4, pages 547–562. University of California Press.
Salat, H., Murcio, R., and Arcaute, E. (2017). Multifractal
methodology. Physica A: Statistical Mechanics and
its Applications, 473:467–487.
Sawano, S., Kodera, S., Setoguchi, N., Tanabe, K., Kushida,
S., Kanda, J., Saji, M., Nanasato, M., Maki, H., Fu-
jita, H., et al. (2024). Applying masked autoencoder-
based self-supervised learning for high-capability vi-
sion transformers of electrocardiographies. Plos one,
19(8):e0307978.
Swapnarekha, H., Nayak, J., Naik, B., and Pelusi, D.
(2024). A deep insight into intelligent fractal-based
image analysis with pattern recognition. In Intelligent
Fractal-Based Image Analysis, pages 3–32. Elsevier.
Yang, J., Shi, R., Wei, D., Liu, Z., Zhao, L., Ke, B., Pfis-
ter, H., and Ni, B. (2023). Medmnist v2-a large-scale
lightweight benchmark for 2d and 3d biomedical im-
age classification. Scientific Data, 10(1):41.
Yang, X., He, X., Zhao, J., Zhang, Y., Zhang, S., and Xie,
P. (2020). Covid-ct-dataset: a ct scan dataset about
covid-19. arXiv preprint arXiv:2003.13865.
Yu, F., Liu, J., Shang, L., and Liu, D. (2022). Multifractal
analysis and stacked autoencoder-based feature learn-
ing method for multivariate processes monitoring. In
2022 41st Chinese Control Conference (CCC), pages
4185–4190. IEEE.
Zhang, Q., Wang, Y., and Wang, Y. (2022). How mask mat-
ters: Towards theoretical understandings of masked
autoencoders. Advances in Neural Information Pro-
cessing Systems, 35:27127–27139.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
776
