
Improved Binary Elk Herd Optimizer with Fitness Balance Distance for
Feature Selection Using Gene Expression Data
Mohamed Wajdi Ouertani
1 a
, Raja Oueslati
1 b
and Ghaith Manita
1,2 c
1
Laboratory MARS, LR17ES05, ISITCom, Sousse University, Sousse, Tunisia
2
ESEN, Manouba University, Manouba, Tunisia
Keywords:
Feature Selection, Optimization, Elk Herd Optimiser, Distance Balance Mechanism.
Abstract:
This research paper introduces an enhanced version of the Binary Elk Herd Optimizer (BEHO), integrated with
a Fitness Distance Balance (FDB) mechanism called FDB-BEHO, tailored for high-dimensional optimization
tasks. This study evaluates the performance of FDB-BEHO across multiple gene expression datasets, focusing
on feature selection in bioinformatics—a domain characterized by complex, high-dimensional data. The FDB
mechanism is designed to prevent premature convergence by maintaining an optimal balance between explo-
ration and exploitation, utilizing a diversity measure that adjusts dynamically based on the fitness-distance
correlation among solutions. Comparative analyses demonstrate that FDB-BEHO surpasses traditional meta-
heuristic algorithms in fitness values and classification accuracy and reduces the number of selected fea-
tures, thereby enhancing model simplicity and interpretability. These results validate the effectiveness of
FDB-BEHO in navigating complex solution spaces efficiently and underscore its potential applicability in
other domains requiring robust feature selection capabilities. The study’s findings suggest that incorporating
diversity-enhancing mechanisms like FDB can significantly improve the performance of binary optimization
algorithms, offering promising directions for future research in optimization technology.
1 INTRODUCTION
In medical research, DNA microarray technology has
revolutionized our ability to analyze gene expression
data, enabling the simultaneous observation of thou-
sands of genes in a single experiment. However, this
advancement also presents a significant challenge:
the curse of dimensionality. With such vast amounts
of data, it becomes crucial to identify and select
the most relevant features—genes that significantly
contribute to accurate disease prediction and classi-
fication. Feature selection (FS) methods are pivotal
in addressing this challenge, allowing researchers
to eliminate irrelevant or redundant genes, thereby
enhancing the performance of predictive models and
reducing computational complexity (Zebari et al.,
2020).
In recent years, the importance of FS has been un-
derscored in various studies focused on cancer pre-
a
https://orcid.org/0009-0000-6164-0069
b
https://orcid.org/0009-0002-5783-5722
c
https://orcid.org/0000-0003-0782-9658
diction (Haq et al., 2021), where identifying key ge-
netic markers is essential for early diagnosis and treat-
ment planning. Traditional FS methods, such as fil-
ter, wrapper, and embedded approaches, have been
widely applied, each with its strengths and limitations
(Venkatesh and Anuradha, 2019). Filters are inde-
pendent of the learning algorithm but may overlook
interactions between features (Bommert et al., 2022);
wrappers are more accurate but computationally ex-
pensive (Maldonado and Weber, 2009); and embed-
ded methods integrate FS within the model training
process, offering a balanced approach (Wang et al.,
2015).
Given the complexity and non-linearity of gene
expression data, metaheuristic algorithms have
emerged as powerful tools for FS (Dokeroglu et al.,
2022). Inspired by natural, biological, or social pro-
cesses, these optimization approaches are designed to
solve complex problems efficiently (Ouertani et al.,
2022a; Oueslati et al., 2024). Metaheuristics excels
at exploring vast search spaces and avoiding subop-
timal solutions, unlike traditional optimization meth-
ods, which may be constrained by linearity or con-
tinuity requirements (Ouertani et al., 2022b). Com-
786
Ouertani, M. W., Oueslati, R. and Manita, G.
Improved Binary Elk Herd Optimizer with Fitness Balance Distance for Feature Selection Using Gene Expression Data.
DOI: 10.5220/0013367600003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 2, pages 786-797
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

mon metaheuristics include evolutionary algorithms
(Srinivas and Patnaik, 1994) such as Artificial Bee
Colony (ABC) (Karaboga and Basturk, 2007), Ge-
netic Algorithm (GA) (Holland, 1992), and Differ-
ential Evolution (DE) (Qin et al., 2008). Swarm-
based optimization methods such as Elk Herd Opti-
mizer (EHO) (Al-Betar et al., 2024), Particle Swarm
Optimization (PSO) (Kennedy and Eberhart, 1995),
and Social Spider Algorithm (SSA) (Mirjalili et al.,
2017) have proven effective in various applications.
Additionally, human-inspired metaheuristics such as
Teaching-Learning Based Optimization (TLBO) (Rao
et al., 2011), Soccer League Competition (SLC)
(Moosavian and Roodsari, 2014), and Brain Storm
Optimization (BSO) (Shi, 2011) leverage social be-
haviors and cognitive processes for optimization. Fi-
nally, physics-based metaheuristics such as Simulated
Annealing (SA) (Bertsimas and Tsitsiklis, 1993),
Atom Search Optimization (ASO) (Zhao et al., 2019),
and Equilibrium Optimizer (EO) (Faramarzi et al.,
2020) draw inspiration from physical phenomena like
thermodynamics and gravitational forces to guide the
search for optimal solutions. Their primary advan-
tages lie in their flexibility and adaptability, making
them suitable for various problems, particularly com-
binatorial optimization challenges. However, they can
be computationally intensive and do not guarantee
globally optimal solutions, often requiring careful pa-
rameter tuning to balance solution quality and com-
putational cost (Nssibi et al., 2024).
In order to address the inherent challenges of
gene expression data analysis (Nssibi et al., 2023)
and as the field continues to evolve, the application
of advanced metaheuristics in FS not only enhances
model accuracy but also offers valuable insights into
complex underlying processes, paving the way for
more targeted and effective decision-making strate-
gies across various domains in bioinformatics (Saeys
et al., 2007). This study introduces an enhanced
version of the Binary Elk Herd Optimizer (BEHO),
named FDB-BEHO, which incorporates the Fitness
Distance Balance (FDB) mechanism to overcome pre-
mature convergence and maintain an optimal balance
between exploration and exploitation. The proposed
algorithm is specifically designed to minimize the
number of selected features, making it well-suited for
addressing high-dimensional optimization challenges
effectively. To evaluate its effectiveness, FDB-BEHO
is tested on nine benchmark biological datasets for
feature selection. Its performance is compared against
state-of-the-art metaheuristic algorithms using met-
rics such as fitness values, classification accuracy, and
feature selection efficiency.
The main objectives and contributions of this
work are as follows:
• Introduction of the Binary EHO (BEHO): A novel
adaptation of the EHO algorithm tailored for fea-
ture selection problems. This binary variant en-
ables the direct application of EHO in solving dis-
crete optimization challenges associated with FS.
• Improvement of BEHO with the FDB Mecha-
nism: A further enhancement of BEHO, incorpo-
rating the FDB mechanism to address issues of
premature convergence. This improvement en-
sures a more effective balance between explo-
ration and exploitation. The enhanced algorithm
dynamically adjusts diversity to maintain robust
performance across varying optimization land-
scapes.
• Evaluation of FDB-BEHO on gene expression
data for FS: Assess the performance of the pro-
posed FDB-BEHO algorithm on nine benchmark
biological datasets for FS. The evaluation involves
a comparative analysis with other state-of-the-art
metaheuristics to validate its efficacy and robust-
ness.
The remainder of this paper is structured as fol-
lows: Section 2 provides a detailed overview of meta-
heuristic optimization in feature selection, highlight-
ing the effectiveness of various algorithms in manag-
ing high-dimensional data. Section 3 introduces the
proposed Binary Elk Herd Optimizer (Binary EHO)
and its enhancement with the Fitness Distance Bal-
ance (FDB) mechanism, including the technical de-
tails of its implementation. Section 4 presents the ex-
perimental setup and the results of applying the pro-
posed method to gene expression datasets, comparing
its performance with other state-of-the-art algorithms.
Finally, Section 5 concludes the paper by discussing
the findings, their implications for feature selection
in bioinformatics, and potential avenues for future re-
search.
2 METAHEURISTIC
OPTIMIZATION IN FEATURE
SELECTION
In this section, we explore the role of metaheuristic
optimization in feature selection, offering a compre-
hensive overview of various algorithms and their suc-
cessful applications in addressing the challenges of
high-dimensional gene expression data.
In (S
¨
onmez et al., 2021), the study explores the use of
hybrid methods combining Genetic Algorithms (GA)
with Support Vector Machines (SVM) and k-Nearest
Improved Binary Elk Herd Optimizer with Fitness Balance Distance for Feature Selection Using Gene Expression Data
787

Neighbors (KNN) for feature selection and classifi-
cation of gene expression datasets. The authors pro-
pose enhancing the GA through the integration of
filter methods such as Pearson’s correlation coeffi-
cient, Relief-F, and mutual information. The study
evaluates these methods across eight gene expres-
sion datasets, primarily related to cancer classifica-
tion. The proposed GA-SVM and GA-KNN meth-
ods demonstrate superior accuracy compared to tra-
ditional approaches, highlighting the effectiveness of
hybrid techniques in reducing the dimensionality of
high-throughput data while maintaining or improv-
ing classification accuracy. The research empha-
sizes the importance of hybrid methods in managing
the computational complexity of large gene expres-
sion datasets and improving predictive performance
in medical applications, particularly in cancer diag-
nosis and subtype classification.
Qin et al. proposed a two-stage feature selection
framework tailored to classify high-dimensional gene
expression data, utilizing an improved Salp Swarm
Algorithm (SSA) (Qin et al., 2022). The first stage
combines Weighted Gene Co-expression Network
Analysis (WGCNA), Random Forest (RF), and Max-
Relevance and Min-Redundancy (mRMR) to initially
reduce the feature space by selecting the most rele-
vant and non-redundant genes. In the second stage,
the improved binary SSA is employed to refine the
feature set further, ensuring a balance between classi-
fication accuracy and the number of selected features.
The framework was tested across ten gene expres-
sion datasets and outperformed other intelligent opti-
mization algorithms like PSO, GWO, and WOA. The
results demonstrated that this method could achieve
high classification accuracy with fewer selected fea-
tures, making it a robust solution for gene expression
data classification in cancer diagnosis.
In (Alzaqebah et al., 2021), Alzaqebah et al. proposed
a Memory-Based Cuckoo Search (MBCS) algorithm
for feature selection in gene expression datasets, par-
ticularly focusing on cancer prediction. The study
addresses the challenges of high-dimensionality and
feature redundancy in microarray data, which can
hinder accurate classification. The MBCS algorithm
enhances the traditional Cuckoo Search Algorithm
(Gandomi et al., 2013) by incorporating a memory
mechanism that records the best solutions found dur-
ing the search process. This memory helps the al-
gorithm avoid re-exploring suboptimal areas and fo-
cuses on promising regions of the search space. The
study tested the algorithm on twelve different mi-
croarray datasets and found that MBCS outperformed
other algorithms, such as Genetic Algorithms (GA),
Particle Swarm Optimization (PSO), and Gravita-
tional Search Algorithm (GSA), in terms of classifi-
cation accuracy and the number of selected features.
The results indicate that MBCS is particularly effec-
tive in reducing the dimensionality of the data while
maintaining or improving the accuracy of cancer pre-
dictions.
Qu et al. introduced an improved Harris Hawks Op-
timization algorithm, tailored explicitly for feature
selection in gene expression data, called the Vari-
able Neighborhood Learning Harris Hawks Optimizer
(VNLHHO) (Qu et al., 2021). This approach was
designed to enhance the global exploration and lo-
cal exploitation capabilities of the standard Harris
Hawks Optimizer (HHO). The VNLHHO incorpo-
rates a dynamic neighborhood learning strategy and
mutation operations to increase population diversity
and prevent the algorithm from falling into local op-
tima. The effectiveness of VNLHHO was validated
across eight cancer gene expression datasets, demon-
strating superior classification accuracy and conver-
gence speed compared to traditional algorithms like
PSO, GA, and the original HHO. The results showed
that VNLHHO not only improved classification per-
formance but also effectively reduced the number of
selected features, making it a powerful tool for high-
dimensional biomedical data analysis.
3 PROPOSED APPROACH
This section presents the proposed approach, which
utilizes the Elk Herd Optimizer (EHO) (Al-Betar
et al., 2024) as the core algorithm for feature selec-
tion in high-dimensional gene expression data. The
approach is systematically organized into three main
components: (i) Overview of EHO: We begin by in-
troducing the EHO algorithm and highlighting its dis-
tinctive characteristics that make it suitable for com-
plex optimization tasks, (ii) Transformation to Binary
EHO: Next, we describe the adaptation of EHO into
a binary format (Binary EHO), specifically tailored
to meet the unique demands of feature selection prob-
lems, and (iii) Integration of Fitness Distance Balance
(FDB): Finally, we incorporate the Fitness Distance
Balance (FDB) mechanism into Binary EHO, signif-
icantly enhancing its capability to maintain a robust
balance between exploration and exploitation while
identifying optimal feature subsets. This structured
approach ensures a comprehensive framework for ad-
dressing the challenges of feature selection in high-
dimensional datasets.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
788

3.1 Elk Herd Optimizer Overview
The Elk Herd Optimizer (EHO) is a novel metaheuris-
tic algorithm inspired by the breeding behavior of elk
herds. It mimics the seasonal dynamics within the
herd, where more muscular bulls lead larger groups
during the rutting season, and new solutions are gen-
erated during the calving season. This approach bal-
ances exploration and exploitation in optimization
tasks, making EHO an effective tool for solving com-
plex problems. Its design is particularly suited for
navigating challenging search spaces and finding op-
timal solutions efficiently. The EHO is designed to
simulate the natural dynamics of elk herds through a
sequence of critical phases. It begins with the initial-
ization of the population and the problem parameters.
The algorithm then enters the rutting season, dividing
the population into families led by the fittest bulls. In
the calving season, these families produce new solu-
tions based on the characteristics of the bull and its
harems. Finally, during the selection season, all so-
lutions are evaluated, and the fittest are selected to
form the next generation, with this process repeating
until the algorithm converges or the iteration limit is
reached. The steps of the EHO are as follows :
1. Initialization: During the initialization phase of
the EHO, the algorithm begins by setting up the
population and defining the problem-specific pa-
rameters. The primary elements to initialize are
the elk herd size (EHS), the bull rate (Br), and the
search space boundaries. The elk herd EH is ini-
tialized as a matrix of size n ×EHS, where n is the
problem’s dimensionality, and each element in the
matrix represents a potential solution (elk). Math-
ematically, each solution x
j
in the population is
generated within the defined search space bound-
aries using Equation 1:
x
i
j
= lb
i
+ (ub
i
− lb
i
) ×U(0, 1) (1)
where x
i
j
represents the i-th attribute of the j-th so-
lution, lb
i
and ub
i
are the lower and upper bounds
for the i-th attribute, and U(0, 1) is a uniformly
distributed random number between 0 and 1. The
fitness of each solution is then calculated using
the objective function f (x), and the solutions are
sorted in ascending order based on their fitness
values. This initial setup prepares the elk herd for
the subsequent phases of the algorithm.
2. Generating the initial Elk Herd Solutions: In the
second step, the algorithm focuses on creating the
initial solutions population, representing the elk
herd. After defining the problem-specific param-
eters and initializing the population matrix EH in
the first step, this phase involves assigning fitness
values to each solution and organizing the herd
structure. The elk herd EH is generated as a ma-
trix of size n × EHS, where each row corresponds
to a potential solution in the search space as pre-
sented in Equation 2.
EH =
x
1
1
x
1
2
··· x
1
n
x
2
1
x
2
2
··· x
2
n
.
.
.
.
.
. ···
.
.
.
x
EHS
1
x
EHS
2
··· x
EHS
n
(2)
Once the initial population is generated, the fit-
ness of each solution is evaluated using the ob-
jective function f (x). The herd is then sorted in
ascending order of fitness, ensuring that the best
solutions (strongest elks) are positioned at the top.
This ordered structure sets the foundation for the
subsequent rutting season phase, where the popu-
lation will be divided into families.
3. Rutting season: In the third step, the EHO algo-
rithm divides the initial population into families,
with each family led by a bull (the fittest indi-
vidual). The division is based on the fitness of
the bulls, reflecting the natural behavior where
stronger bulls lead larger groups. First, the al-
gorithm determines the number of bulls B in the
population using the bull rate Br and the elk herd
size EHS as shown in Equation 3. :
B = Br × EHS (3)
where B is the number of bulls, Br is the bull rate,
and EHS is the total population size. The top B
individuals, based on their fitness values, are se-
lected as bulls. Next, the bulls compete to form
families, each consisting of a bull and its assigned
harems (followers). The assignment of harems to
each bull is done using a roulette-wheel selection
process, where the probability p
j
of a bull j at-
tracting a harem is proportional to its fitness pre-
sented in Equation 4:
p
j
=
f (x
j
)
∑
B
k=1
f (x
k
)
(4)
Here, f (x
j
) is the fitness of the j-th bull, and the
sum in the denominator is the total fitness of all
bulls. The roulette-wheel selection ensures that
bulls with higher fitness are more likely to lead
more harems. Once the harems are assigned, each
bull leads its family, with the size of each fam-
ily reflecting the strength of the bull. This struc-
tured division sets the stage for the calving season,
where new solutions (calves) will be generated
based on the bulls’ characteristics and harems.
Improved Binary Elk Herd Optimizer with Fitness Balance Distance for Feature Selection Using Gene Expression Data
789
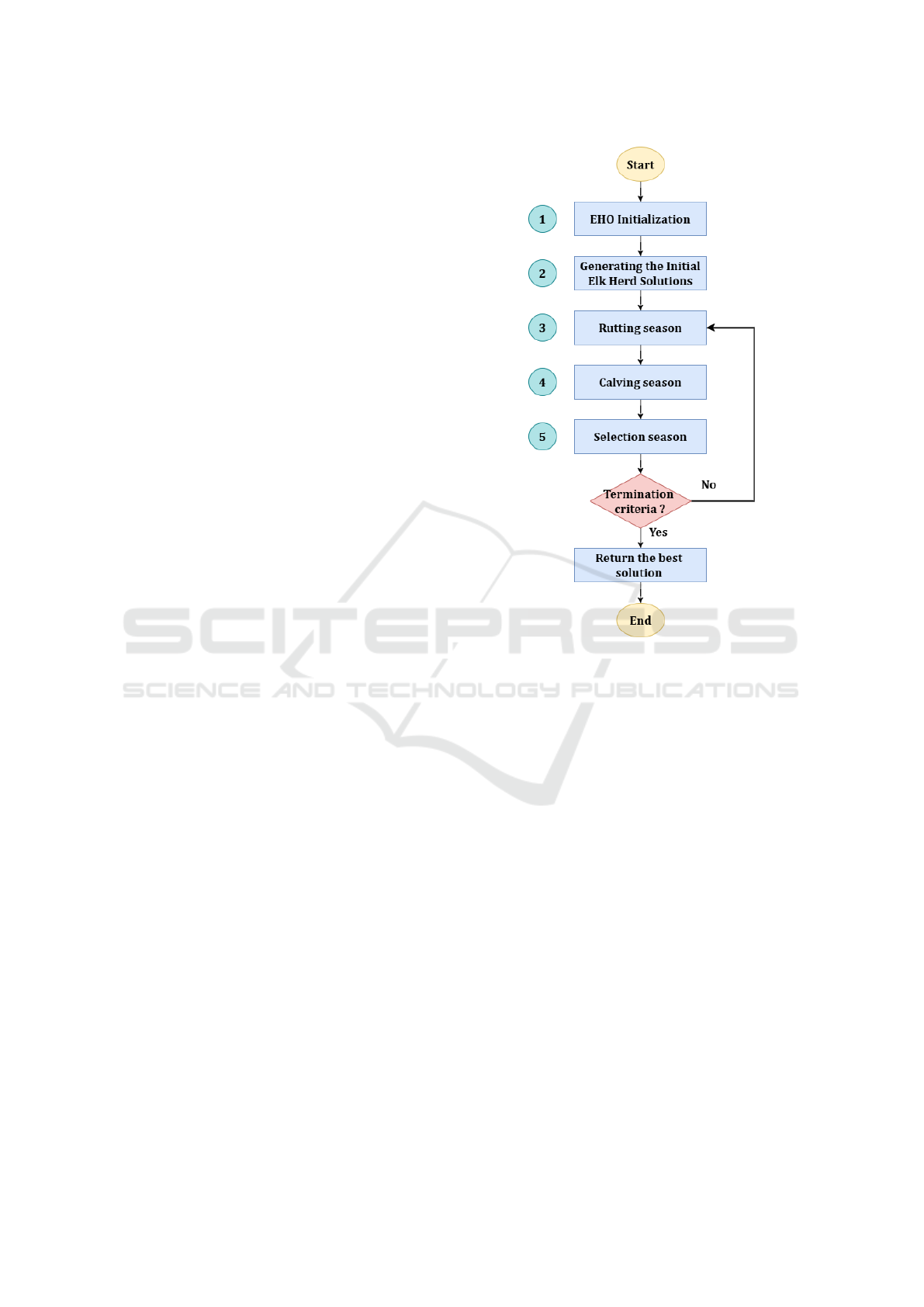
4. Calving season: In the fourth step, the EHO
algorithm focuses on generating new solutions
(calves) within each family based on the genetic
traits of the bull (leader) and its harems (follow-
ers). This process mimics the natural reproduc-
tion process in elk herds, where the offspring in-
herit characteristics from both parents, promoting
diversity within the population. For each family,
new solutions x
i
j
(t + 1) are generated by combin-
ing attributes from the bull x
j
and its harems x
i
j
(t).
If the calf’s index matches that of its bull father,
the new solution is generated using Equation 5:
x
i
j
(t + 1) = x
i
j
(t) + α · (x
i
k
(t) − x
i
j
(t)) (5)
where α is a random number between 0 and 1,
and x
i
k
(t) is a randomly selected attribute from
the current population. This equation ensures that
the new solution is influenced primarily by the
bull, with some variation introduced by the ran-
dom selection from the herd. If the calf’s index
matches that of its mother harem, the new solu-
tion is created by combining the attributes of both
the mother and the bull, using Equation 6:
x
i
j
(t +1) = x
i
j
(t)+β·(x
i
h j
(t)−x
i
j
(t))+γ·(x
i
r
(t)−x
i
j
(t))
(6)
Here, β and γ are random numbers in the range
[0, 2], x
i
h j
(t) represents the bull’s attributes, and
x
i
r
(t) is a random attribute from another bull. This
equation allows the calf to inherit traits from both
parents, with additional diversity introduced by
the random selection. This calving process con-
tinues for all families, producing a new genera-
tion of solutions that inherit their predecessors’
strengths while introducing new variations, which
is crucial for the algorithm’s exploration and ex-
ploitation capabilities in the search space.
5. Selection season: In the fifth step, the EHO al-
gorithm consolidates the newly generated solu-
tions (calves) with the existing population of bulls
and harems to form a unified herd. This phase
aims to evaluate the fitness of all individuals in
this combined population and select the best solu-
tions to carry forward to the next generation. First,
the bulls, harems, and newly generated calves are
merged into a single matrix, EH
temp
. Each indi-
vidual’s fitness in EH
temp
is evaluated using the
objective function, and the entire population is
sorted in ascending order based on fitness values.
From this sorted population, the top EHS indi-
viduals, where EHS is the elk herd size, are se-
lected to form the new population for the next iter-
ation. This selection process ensures that only the
fittest individuals, whether they are bulls, harems,
or calves, are retained in the herd. This method
Figure 1: Flowchart of the Elk Herd Optimizer.
of selection is akin to the µ + λ selection strat-
egy commonly used in evolutionary algorithms,
where both parents (bulls and harems) and off-
spring (calves) compete equally for survival. By
continuously selecting the fittest individuals, the
algorithm iteratively refines the population, im-
proving the overall fitness of the herd with each
cycle. This selection process repeats until the ter-
mination criteria, such as a maximum number of
iterations or convergence to an optimal solution,
are met.
The flowchart and pseudocode of the EHO are pre-
sented in Figure 1 and Algorithm 1, respectively.
3.2 Binary Elk Herd Optimizer
Transforming the EHO into a Binary EHO is essential
for adapting the algorithm to feature selection tasks,
which require decisions to be made in a binary search
space. In feature selection, each candidate solution
is represented as a binary vector where each element
indicates whether a particular feature is included (1)
or excluded (0). The transformation of EHO to handle
binary vectors involves the following technical steps:
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
790

Algorithm 1: Elk Herd Optimizer (EHO) Algorithm.
1: Input: Population size (EHS), Bull rate (Br),
Maximum iterations (MaxIter)
2: Output: Best solution found
3: Initialize population EH with EHS solutions ran-
domly within the search space
4: Evaluate the fitness of each solution in EH
5: Sort EH based on fitness in ascending order
6: for iter = 1 to MaxIter do
7: Determine number of bulls B = ⌈Br × EHS⌉
8: Select the top B solutions as bulls from EH
9: Assign harems to each bull using roulette-
wheel selection based on fitness
10: for each family (bull and its harems) do
11: if index matches bull then
12: x
new
= x
bull
+ α × (x
random
− x
bull
)
13: else
14: x
new
= x
harem
+ β × (x
bull
− x
harem
) +
γ × (x
random bull
− x
harem
)
15: end if
16: end for
17: Combine bulls, harems, and calves into
EH
temp
18: Evaluate fitness of all solutions in EH
temp
19: Sort EH
temp
based on fitness in ascending or-
der
20: Select the top EHS solutions from EH
temp
to
form the new population EH
21: end for
22: Return the best solution in EH
1. Binary Representation: Instead of representing
solutions as continuous vectors, each solution is
now a binary vector x = [x
1
, x
2
, . . . , x
n
], where
x
i
∈ {0, 1} for each feature i. This change allows
the algorithm to directly address feature selection
by determining whether each feature should be in-
cluded in the model.
2. Position Update with Transfer Functions: The
core challenge in adapting EHO to a binary for-
mat lies in converting the continuous position up-
dates, typical in EHO, to binary updates. This is
achieved using transfer functions (Equations 7,8)
(Mirjalili and Lewis, 2013; Nssibi et al., 2021).
After the continuous update equation is computed
for each element x
i
j
of the solution vector, a trans-
fer function T F(x) is applied to convert this con-
tinuous value into a probability. Common transfer
functions include:
• Sigmoid Function (S-shaped):
T F(x) =
1
1 + e
−x
(7)
• V-shaped Function:
T F(x) = |tanh(x)| (8)
The output of these functions lies between 0 and
1 and represents the probability that a particular
feature will switch its state (from 0 to 1 or vice
versa).
3. Binary Decision-Making: Once the probability
P(x
i
j
) is determined using the transfer function,
the next step is to convert this probability into a
binary decision. This is done by comparing the
probability with a random number rand() gener-
ated uniformly between 0 and 1 as presented in
Equation 9:
x
i
j
=
(
1, if P(x
i
j
) ≥ rand()
0, otherwise
(9)
This comparison ensures that each feature’s bi-
nary state is updated based on the likelihood cal-
culated through the transfer function.
4. Fitness Evaluation: After updating the binary vec-
tors, the fitness of each solution is evaluated. The
fitness function typically balances the trade-off
between maximizing classification accuracy (us-
ing a classifier like k-NN) and minimizing the
number of features selected. This ensures that the
selected subset is both practical and compact.
By incorporating these steps, the Binary EHO can
effectively navigate binary search spaces, making it
highly suitable for feature selection tasks. Trans-
fer functions are critical in this transformation, as
they bridge the gap between continuous optimization
strategies and the discrete nature of feature selection
problems, enabling the algorithm to retain its explo-
ration and exploitation capabilities in a binary con-
text.
3.3 Fitness Distance Balance BEHO
Integrating the Fitness Distance Balance (FDB)
(Kahraman et al., 2020) mechanism into the BEHO
enhances its performance in feature selection by in-
troducing a balance between exploration and exploita-
tion. This mechanism evaluates each solution based
not only on its fitness but also on its diversity rela-
tive to the best-known solution. Technically, this is
achieved by first calculating the Euclidean distance
D
j
between each binary solution x
j
and the current
best solution Best
current
, using the formula in Equa-
tion 10:
D
j
=
s
n
∑
i=1
(x
i
j
− Best
i
current
)
2
(10)
Improved Binary Elk Herd Optimizer with Fitness Balance Distance for Feature Selection Using Gene Expression Data
791
Next, both the fitness values and the distances are nor-
malized to ensure no single metric dominates as pre-
sented in Equations 11 and 12 :
F
norm
( j) =
F( j) − min(F)
max(F) − min(F)
(11)
D
norm
( j) =
D
j
− min(D)
max(D) − min(D)
(12)
These normalized values are then combined into a
composite score S
j
that balances fitness and diversity
as presented in Equation 13:
S
j
= α × F
norm
( j) + (1 − α) × D
norm
( j) (13)
This score guides the selection process, with higher-
scoring solutions more likely to be retained or se-
lected as bulls. Using this composite score, the al-
gorithm avoids premature convergence and maintains
a diverse population, which is crucial for effectively
exploring the binary search space and identifying op-
timal feature subsets. This approach significantly
enhances the robustness and effectiveness of Binary
EHO in feature selection tasks. The complete pseu-
docode of the FDB-BEHO is presented in Algorithm
2.
Algorithm 2: Binary EHO with Fitness Distance Balance
(FDB) Mechanism.
1: Initialize population EH as binary vectors
2: for iter = 1 to MaxIter do
3: Determine bulls and harems using binary fit-
ness and distance evaluation
4: for each family do
5: Calculate continuous updates for posi-
tions
6: Apply transfer function to convert updates
to probabilities
7: Update binary positions using the calcu-
lated probabilities
8: end for
9: Calculate distance D
j
for each solution from
the best solution
10: Normalize fitness and distance values
11: Compute composite score S
j
=
α × F
norm
( j) + (1 − α) × D
norm
( j)
12: Select the best solutions based on composite
scores to form the new population
13: end for
14: Return the best binary solution
4 EXPERIMENTAL RESULTS
In this section, we evaluate the performance of the
proposed FDB-BEHO across various gene expres-
sion datasets. All experiments were conducted us-
ing MATLAB R2020a on an Intel Core i5 machine
with a 3.3 GHz CPU and 12GB of RAM. Each ex-
periment was repeated 30 times to ensure statistical
significance.
4.1 Datasets
We utilized the following nine benchmark bi-
ological datasets to assess the effectiveness of
the FDB-BEHO: Leukemia (Golub et al., 1999),
Prostate GE (Singh et al., 2002), Colon (Alon et al.,
1999), Lung discrete (Bhattacharjee et al., 2001),
SMK CAN 187 (Spira et al., 2007), Lymphoma
(Alizadeh et al., 2000), CLL SUB 111 (Haslinger
et al., 2004), Lung (Bhattacharjee et al., 2001),
and nci9 (Ross et al., 2000). The details of these
datasets, including the number of instances, features,
and classes, are summarized in Table 1. The parame-
ters for FDB-BEHO and the comparative metaheuris-
tic algorithms are detailed in Table 2.
Table 1: Summary of Datasets Used in the Experiments.
Dataset Instances Features Classes
CLL SUB 111 111 11340 3
Colon 62 2000 2
Leukemia 72 7070 2
Lung 203 3312 5
Lung discrete 73 325 7
Lymphoma 96 4026 9
nci9 60 9712 9
Prostate GE 102 5966 2
SMK CAN 187 187 19993 2
Table 2: Parameter Settings for the Algorithms.
Algorithm Parameter Value
FDB-BEHO
EHS ( Population Size)
Br (Break Rate)
10
0.3
SSA c1 (leader position update probability) 0.5
PSO
c1, c2 (acceleration coefficients)
ω (inertia weight)
2
0.1
GA
crossover rate
mutation rate
0.9
0.1
EO
α
β
1
2
ASO
α (depth weight)
β (multiplier weight)
Vmax (maximum velocity)
50
0.2
6
All of them
search agents (bats, wolves, particles,...)
maximum iterations
30
200
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
792

4.2 Results and Discussion
This section thoroughly examines various datasets,
illustrating the performance of the FDB-BEHO op-
timization algorithm variants compared with estab-
lished algorithms such as ABC, SSA, PSO, GA, EO,
and ASO. The metrics for this comparison encom-
pass fitness values, classification accuracy, the num-
ber of selected features, and statistical significance as
assessed by the Wilcoxon test.
In Table 3, the analysis of fitness values shows
FDB-BEHO-V as a standout performer, consistently
achieving the lowest fitness scores on challeng-
ing datasets such as CLL SUB 111, colon, and
leukaemia. These results demonstrate the algorithm’s
superior capability in efficiently navigating complex
solution spaces to identify highly optimal solutions.
Moreover, FDB-BEHO-V exhibits the lowest stan-
dard deviations among the compared algorithms, em-
phasising its stability and reliability. Such consis-
tency is crucial in optimization tasks where depend-
ability and predictability of performance are as cru-
cial as the performance itself.
The narrative continues in Table 4, focusing on
classification accuracy. Here, FDB-BEHO-S fre-
quently outperforms FDB-BEHO-V, indicating its en-
hanced capability in models where higher accuracy
is paramount. However, the performance landscape
is nuanced, with algorithms like EO and PSO ex-
celling in specific datasets, particularly lung and
Prostate GE. These observations underline the neces-
sity of adaptive algorithm selection based on specific
dataset characteristics, suggesting that no single al-
gorithm uniformly outperforms others across all con-
texts. This variability also highlights the importance
of understanding the underlying data characteristics
and choosing algorithms that align well with those
characteristics to optimize performance.
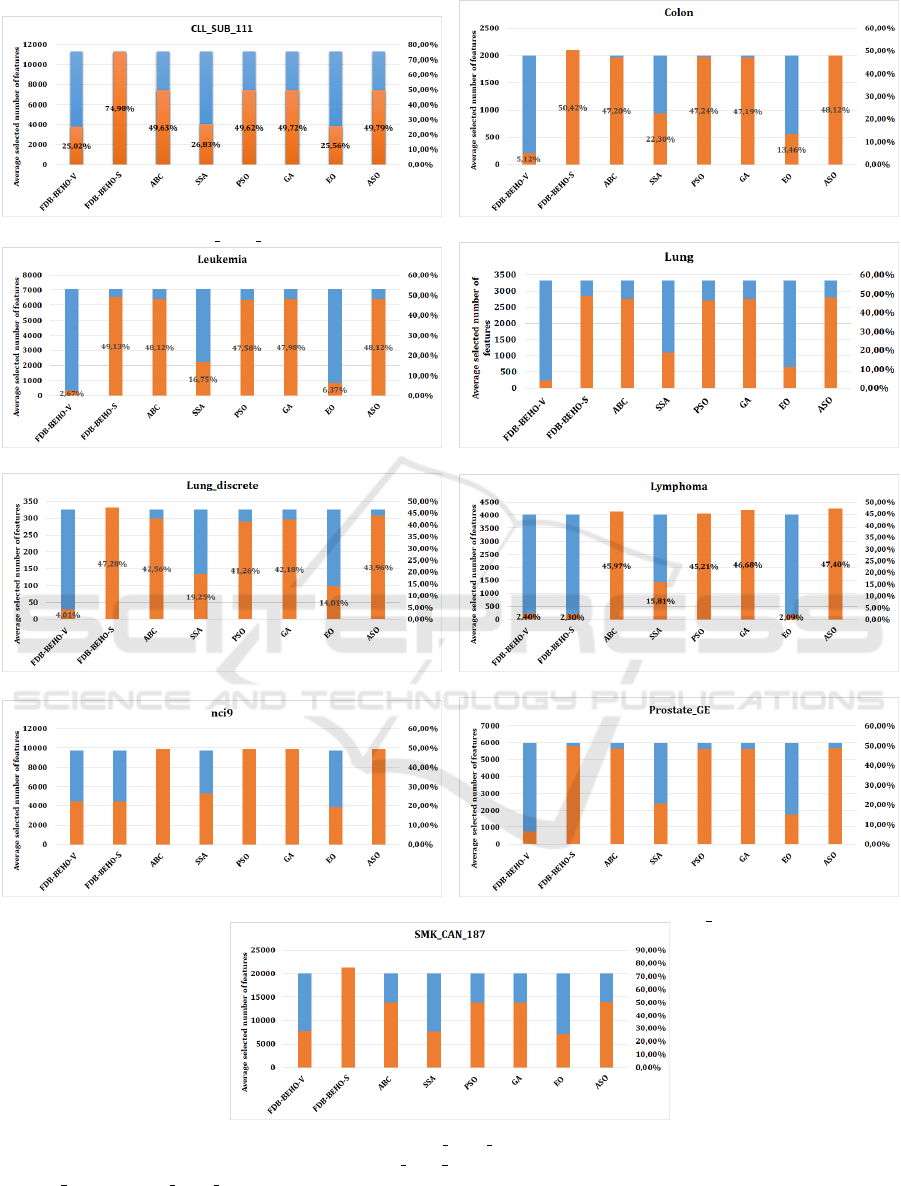
Table 5 sheds light on the efficiency of feature
selection, a critical metric that affects both the com-
plexity and the computational efficiency of the result-
ing models. FDB-BEHO-V is particularly adept at
reducing the number of features required to achieve
high performance, which is evident in its handling
of datasets like CLL SUB 111 and colon. By se-
lecting fewer features, FDB-BEHO-V not only sim-
plifies the complexity of the models but also po-
tentially enhances the interpretability of the results,
which is invaluable in applications where understand-
ing the algorithm’s decision-making process is essen-
tial. Moreover, models with fewer features generally
train and deploy faster, offering practical advantages
in real-time applications. Hence, Figure 2 provides a
clearer visualization of the obtained results.
The robustness of these results is further validated
in Table 6, where the Wilcoxon test results confirm
the statistical significance of the improvements of-
fered by FDB-BEHO-V over other algorithms in most
datasets. However, in datasets like lymphoma and
nci9, where no significant differences are found, the
results suggest that FDB-BEHO-V may not always
offer a decisive advantage, indicating potential areas
for further algorithmic refinement and improvement.
This detailed analysis underscores FDB-BEHO-
V’s capabilities to deliver top-tier optimization per-
formance with robust reliability across a range of
domains, presenting it as an excellent candidate for
tackling complex optimization problems in varied set-
tings. The findings from this investigation are crucial
for advancing the development and application of op-
timization methodologies in both academic research
and practical industrial applications, driving innova-
tion and efficiency in this vital field.
5 CONCLUSION
This study has meticulously explored the enhance-
ments to the BEHO by integrating the Fitness Dis-
tance Balance (FDB) mechanism. The adapted
BEHO has been rigorously tested across various gene
expression datasets, demonstrating its ability to effi-
ciently and reliably identify optimal solutions. Intro-
ducing a binary framework tailored for discrete op-
timization tasks such as feature selection has shown
significant promise, particularly in bioinformatics,
where the dimensionality and complexity of data of-
ten pose substantial challenges.
The FDB-BEHO variant has consistently outper-
formed traditional metaheuristic algorithms in terms
of fitness values, classification accuracy, and feature
selection efficiency. This robust performance under-
scores the algorithm’s refined balance between explo-
ration and exploitation, facilitated by the FDB mecha-
nism, which integrates a diversity measure to prevent
premature convergence. The findings from this study
advocate for the potential of BEHO in bioinformatics
and highlight its adaptability and efficiency in manag-
ing high-dimensional datasets.
Looking forward, optimization in high-
dimensional data analysis presents several avenues
for further research. BEHO’s adaptability could be
explored in other complex optimization scenarios
beyond bioinformatics, such as in finance, robotics,
and climate modeling, where similar challenges re-
garding high dimensionality and feature redundancy
exist. Future studies could also delve into hybrid ap-
proaches combining BEHO with other metaheuristic
Improved Binary Elk Herd Optimizer with Fitness Balance Distance for Feature Selection Using Gene Expression Data
793
(a) CLL SUB 111 (b) Colon
(c) Leukemia (d) Lung
(e) Lung discrete (f) Lymphoma
(g) nci9 (h) Prostate GE
(i) SMK CAN 187
Figure 2: Average selected number of features for CLL SUB 111, Colon, Leukemia, Lung, Lung discrete, Lymphoma, nci9,
Prostate GE, and SMK CAN 187 datasets.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
794

Table 3: Comparison of FDB-BEHO versus other optimization algorithms in terms of fitness values.
Dataset Metric FDB- FDB- ABC SSA PSO GA EO ASO
BEHO-V BEHO-S
CLL SUB 111 Best 0,0469 0,0524 0,0948 0,0472 0,0500 0,0499 0,0914 0,0949
Avg 0,0872 0,1019 0,1646 0,1526 0,1505 0,1458 0,1410 0,1585
Std 0,0169 0,0223 0,0302 0,0308 0,0387 0,0310 0,0294 0,0326
colon Best 0,0002 0,0046 0,0045 0,0014 0,0044 0,0045 0,0006 0,0045
Avg 0,0005 0,0050 0,0047 0,0022 0,0047 0,0047 0,0013 0,0048
Std 0,0001 0,0004 7,64E-05 0,0003 0,0001 5,18E-05 0,0003 9,09E-05
leukemia Best 0,0001 0,0048 0,0047 0,0014 0,0046 0,0047 0,0002 0,0047
Avg 0,0002 0,0049 0,0048 0,0016 0,0047 0,0047 0,0006 0,0048
Std 6,63E-05 4,03E-05 3,88E-05 0,0001 4,99E-05 2,30E-05 0,0002 5,37E-05
lung Best 0,0002 0,0047 0,0045 0,0013 0,0044 0,0045 0,0005 0,0046
Avg 0,0003 0,0048 0,0047 0,0018 0,0046 0,0047 0,0011 0,0047
Std 0,0001 5,55E-05 5,67E-05 0,0002 7,69E-05 3,39E-05 0,0002 5,64E-05
lung discrete Best 0,0002 0,0042 0,0039 0,0012 0,0035 0,0038 0,0008 0,0039
Avg 0,0004 0,0047 0,0042 0,0019 0,0041 0,0042 0,0014 0,0043
Std 8,76E-05 0,0002 0,0001 0,0002 0,0002 0,0001 0,0003 0,0002
lymphoma Best 0,0521 0,05222 0,1086 0,1056 0,1086 0,1087 0,0522 0,1088
Avg 0,0543 0,09718 0,1088 0,1057 0,1087 0,1088 0,0901 0,1089
Std 0,0101 0,01807 6,22E-05 7,55E-05 6,83E-05 3,23E-05 0,0234 5,15E-05
nci9 Best 0,0845 0,1651 0,0875 0,0861 0,1698 0,0874 0,0843 0,1698
Avg 0,1478 0,2244 0,2314 0,2161 0,2217 0,2087 0,2203 0,2120
Std 0,0348 0,03755 0,0398 0,0342 0,0470 0,0402 0,0429 0,0461
Prostate GE Best 0,0003 0,0048 0,0047 0,0015 0,0046 0,0047 0,0006 0,0047
Avg 0,0006 0,0049 0,0048 0,0020 0,0048 0,0048 0,0015 0,0048
Std 0,0001 5,71E-05 4,05E-05 0,0003 5,40E-05 3,54E-05 0,0004 7,09E-05
SMK CAN 187 Best 0,0555 0,0348 0,0851 0,0554 0,1119 0,0852 0,0827 0,0852
Avg 0,0961 0,1010 0,1419 0,1417 0,1413 0,1326 0,1394 0,1466
Std 0,0170 0,0179 0,0182 0,0233 0,0201 0,0185 0,0241 0,0202
Table 4: Comparison of FDB-BEHO versus other optimization algorithms in terms of average classification accuracy.
Dataset FDB-BEHO-V FDB-BEHO-S ABC SSA PSO GA EO ASO
CLL SUB 111 0,5606 0,5891 0,5517 0,5294 0,5267 0,5303 0,5472 0,5544
colon 0,8023 0,8137 0,7745 0,7418 0,7778 0,7794 0,7418 0,7696
leukemia 0,895 0,909 0,888 0,8641 0,8613 0,8711 0,8711 0,8838
lung 0,9358 0,948 0,9529 0,948 0,948 0,9515 0,9554 0,9583
lung discrete 0,7969 0,7661 0,8347 0,8039 0,8515 0,8347 0,8417 0,8389
lymphoma 0,8369 0,8710 0,8638 0,8503 0,8648 0,8555 0,8689 0,8596
nci9 0,4003 0,4134 0,4003 0,40458 0,4003 0,3905 0,4003 0,4003
Prostate GE 0,849 0,8265 0,8412 0,8569 0,8667 0,8716 0,8275 0,8343
SMK CAN 187 0,6571 0,6412 0,6402 0,6423 0,6439 0,6471 0,6767 0,6449
or machine learning methods to enhance its efficiency
and effectiveness further.
Moreover, the impact of different parameter set-
tings on BEHO’s performance could be subjected
to a more granular analysis to optimize its applica-
tion across various domains. Integrating advanced
machine learning techniques, such as deep learning,
within the BEHO framework could provide a deeper
understanding of the data structures and feature inter-
actions, potentially leading to more innovative solu-
tions and applications.
Additionally, parallel versions of BEHO could be
developed to leverage modern computational archi-
tectures, significantly reducing the time required for
large-scale computations and making the algorithm
more practical for real-world applications that de-
mand rapid processing speeds.
As the field of optimization continues to evolve,
BEHO’s flexibility and robustness, particularly in its
binary incarnation with the FDB mechanism, position
it as a potent tool capable of making significant con-
tributions to various scientific and engineering dis-
ciplines. The insights gained from this study pave
the way for more targeted and effective optimization
strategies, facilitating advancements in both theoreti-
cal research and practical applications.
Improved Binary Elk Herd Optimizer with Fitness Balance Distance for Feature Selection Using Gene Expression Data
795

Table 5: Comparison of FDB-BEHO versus other optimization algorithms in terms of average selected number of features.
Dataset FDB-BEHO-V FDB-BEHO-S ABC SSA PSO GA EO ASO
CLL SUB 111 2837,16 8502,69 5627,75 3042,80 5626,82 5638,17 2898,14 5645,71
colon 102,41 1008,31 943,96 446,08 944,82 943,82 269,14 962,33
leukemia 189,06 3473,45 3402,10 1184,12 3364,12 3391,89 450,22 3440,25
lung 130,45 1605,61 1561,96 626,45 1536,14 1558,65 366,61 1589,00
lung discrete 13,02 153,67 138,33 62,55 134,08 137,07 45,53 142,86
lymphoma 96,43 92,48 1850,75 636,55 1820,10 1879,19 84,02 1908,35
nci9 2172,55 2167,76 4797,90 2561,59 4800,18 4799,96 1876,92 4806,20
Prostate GE 387,94 2952,61 2890,92 1231,06 2878,24 2885,34 900,16 2919,59
SMK CAN 187 5522,69 15338,35 9969,24 5502,22 9944,98 9964,37 5124,49 10021,29
Table 6: p-values of the Wilcoxson test of FDB-BEHO-V versus other optimization algorithms (p ≥ 0.05 are underlined).
Dataset FDB-BEHO-S ABC SSA PSO GA EO ASO
CLL SUB 111 1,83E-06 6,15E-10 2,80E-09 1,77E-09 8,27E-10 1,05E-09 5,14E-10
colon 5,14E-10 5,13E-10 5,14E-10 5,14E-10 5,13E-10 5,46E-10 5,14E-10
leukemia 5,14E-10 5,13E-10 5,14E-10 5,14E-10 5,14E-10 6,53E-10 5,14E-10
lung 5,14E-10 5,14E-10 5,14E-10 5,14E-10 5,13E-10 5,14E-10 5,13E-10
lung discrete 5,04E-10 5,00E-10 5,07E-10 5,09E-10 4,98E-10 5,09E-10 5,08E-10
lymphoma 0,0608 5,14E-10 5,14E-10 5,13E-10 5,13E-10 4,17E-08 5,14E-10
nci9 0,0608 8,77E-10 6,92E-09 5,15E-10 5,15E-10 2,44E-08 5,15E-10
Prostate GE 5,14E-10 5,14E-10 5,14E-10 5,14E-10 5,14E-10 5,79E-10 5,14E-10
SMK CAN 187 6,08E-02 5,15E-10 2,23E-09 5,15E-10 1,05E-09 4,42E-09 6,93E-10
REFERENCES
Al-Betar, M. A., Awadallah, M. A., Braik, M. S., Makhad-
meh, S., and Doush, I. A. (2024). Elk herd optimizer:
a novel nature-inspired metaheuristic algorithm. Arti-
ficial Intelligence Review, 57(3):48.
Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos,
I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran,
T., Yu, X., et al. (2000). Distinct types of diffuse large
b-cell lymphoma identified by gene expression profil-
ing. Nature, 403(6769):503–511.
Alon, U., Barkai, N., Notterman, D. A., Gish, K., Ybarra,
S., Mack, D., and Levine, A. J. (1999). Broad patterns
of gene expression revealed by clustering analysis of
tumor and normal colon tissues probed by oligonu-
cleotide arrays. Proceedings of the National Academy
of Sciences, 96(12):6745–6750.
Alzaqebah, M., Briki, K., Alrefai, N., Brini, S., Jawarneh,
S., Alsmadi, M. K., Mohammad, R. M. A., AL-
marashdeh, I., Alghamdi, F. A., Aldhafferi, N., et al.
(2021). Memory based cuckoo search algorithm for
feature selection of gene expression dataset. Informat-
ics in Medicine Unlocked, 24:100572.
Bertsimas, D. and Tsitsiklis, J. (1993). Simulated anneal-
ing. Statistical science, 8(1):10–15.
Bhattacharjee, A., Richards, W. G., Staunton, J., Li, C.,
Monti, S., Vasa, P., Ladd, C., Beheshti, J., Bueno, R.,
Gillette, M., et al. (2001). Classification of human
lung carcinomas by mrna expression profiling reveals
distinct adenocarcinoma subclasses. Proceedings of
the National Academy of Sciences, 98(24):13790–
13795.
Bommert, A., Welchowski, T., Schmid, M., and Rah-
nenf
¨
uhrer, J. (2022). Benchmark of filter methods
for feature selection in high-dimensional gene ex-
pression survival data. Briefings in Bioinformatics,
23(1):bbab354.
Dokeroglu, T., Deniz, A., and Kiziloz, H. E. (2022). A com-
prehensive survey on recent metaheuristics for feature
selection. Neurocomputing, 494:269–296.
Faramarzi, A., Heidarinejad, M., Stephens, B., and Mir-
jalili, S. (2020). Equilibrium optimizer: A novel
optimization algorithm. Knowledge-based systems,
191:105190.
Gandomi, A. H., Yang, X.-S., and Alavi, A. H. (2013).
Cuckoo search algorithm: a metaheuristic approach
to solve structural optimization problems. Engineer-
ing with computers, 29:17–35.
Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasen-
beek, M., Mesirov, J. P., Coller, H., Loh, M. L., Down-
ing, J. R., Caligiuri, M. A., et al. (1999). Molecu-
lar classification of cancer: class discovery and class
prediction by gene expression monitoring. science,
286(5439):531–537.
Haq, A. U., Li, J. P., Saboor, A., Khan, J., Wali, S., Ahmad,
S., Ali, A., Khan, G. A., and Zhou, W. (2021). Detec-
tion of breast cancer through clinical data using super-
vised and unsupervised feature selection techniques.
IEEE Access, 9:22090–22105.
Haslinger, C., Schweifer, N., Stilgenbauer, S., Dohner, H.,
Lichter, P., Kraut, N., Stratowa, C., and Abseher, R.
(2004). Microarray gene expression profiling of b-cell
chronic lymphocytic leukemia subgroups defined by
genomic aberrations and vh mutation status. Journal
of Clinical Oncology, 22(19):3937–3949.
Holland, J. H. (1992). Genetic algorithms. Scientific amer-
ican, 267(1):66–73.
Kahraman, H. T., Aras, S., and Gedikli, E. (2020). Fitness-
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
796

distance balance (fdb): a new selection method for
meta-heuristic search algorithms. Knowledge-Based
Systems, 190:105169.
Karaboga, D. and Basturk, B. (2007). A powerful and ef-
ficient algorithm for numerical function optimization:
artificial bee colony (abc) algorithm. Journal of global
optimization, 39:459–471.
Kennedy, J. and Eberhart, R. (1995). Particle swarm opti-
mization. In Proceedings of ICNN’95-international
conference on neural networks, volume 4, pages
1942–1948. IEEE.
Maldonado, S. and Weber, R. (2009). A wrapper method
for feature selection using support vector machines.
Information Sciences, 179(13):2208–2217.
Mirjalili, S., Gandomi, A. H., Mirjalili, S. Z., Saremi, S.,
Faris, H., and Mirjalili, S. M. (2017). Salp swarm
algorithm: A bio-inspired optimizer for engineering
design problems. Advances in engineering software,
114:163–191.
Mirjalili, S. and Lewis, A. (2013). S-shaped versus v-
shaped transfer functions for binary particle swarm
optimization. Swarm and Evolutionary Computation,
9:1–14.
Moosavian, N. and Roodsari, B. K. (2014). Soccer league
competition algorithm: A novel meta-heuristic algo-
rithm for optimal design of water distribution net-
works. Swarm and Evolutionary Computation, 17:14–
24.
Nssibi, M., Manita, G., Chhabra, A., Mirjalili, S., and Ko-
rbaa, O. (2024). Gene selection for high dimensional
biological datasets using hybrid island binary artificial
bee colony with chaos game optimization. Artificial
Intelligence Review, 57(3):51.
Nssibi, M., Manita, G., and Korbaa, O. (2021). Binary giza
pyramids construction for feature selection. Procedia
Computer Science, 192:676–687.
Nssibi, M., Manita, G., and Korbaa, O. (2023). Advances in
nature-inspired metaheuristic optimization for feature
selection problem: A comprehensive survey. Com-
puter Science Review, 49:100559.
Ouertani, M. W., Manita, G., and Korbaa, O. (2022a). Au-
tomatic data clustering using hybrid chaos game opti-
mization with particle swarm optimization algorithm.
Procedia Computer Science, 207:2677–2687.
Ouertani, M. W., Manita, G., and Korbaa, O. (2022b). Im-
proved antlion algorithm for electric vehicle charging
station placement. In 2022 IEEE 9th International
Conference on Sciences of Electronics, Technolo-
gies of Information and Telecommunications (SETIT),
pages 265–271. IEEE.
Oueslati, R., Manita, G., Chhabra, A., and Korbaa, O.
(2024). Chaos game optimization: A comprehensive
study of its variants, applications, and future direc-
tions. Computer Science Review, 53:100647.
Qin, A. K., Huang, V. L., and Suganthan, P. N. (2008). Dif-
ferential evolution algorithm with strategy adaptation
for global numerical optimization. IEEE transactions
on Evolutionary Computation, 13(2):398–417.
Qin, X., Zhang, S., Yin, D., Chen, D., and Dong, X. (2022).
Two-stage feature selection for classification of gene
expression data based on an improved salp swarm al-
gorithm. Math. Biosci. Eng, 19(12):13747–13781.
Qu, C., Zhang, L., Li, J., Deng, F., Tang, Y., Zeng, X., and
Peng, X. (2021). Improving feature selection perfor-
mance for classification of gene expression data using
harris hawks optimizer with variable neighborhood
learning. Briefings in bioinformatics, 22(5):bbab097.
Rao, R. V., Savsani, V. J., and Vakharia, D. (2011).
Teaching–learning-based optimization: a novel
method for constrained mechanical design op-
timization problems. Computer-aided design,
43(3):303–315.
Ross, D. T., Scherf, U., Eisen, M. B., Perou, C. M., Rees,
C., Spellman, P., Iyer, V., Jeffrey, S. S., Van de Rijn,
M., Waltham, M., et al. (2000). Systematic variation
in gene expression patterns in human cancer cell lines.
Nature genetics, 24(3):227–235.
Saeys, Y., Inza, I., and Larranaga, P. (2007). A review of
feature selection techniques in bioinformatics. bioin-
formatics, 23(19):2507–2517.
Shi, Y. (2011). Brain storm optimization algorithm. In Ad-
vances in Swarm Intelligence: Second International
Conference, ICSI 2011, Chongqing, China, June 12-
15, 2011, Proceedings, Part I 2, pages 303–309.
Springer.
Singh, D., Febbo, P. G., Ross, K., Jackson, D. G., Manola,
J., Ladd, C., Tamayo, P., Renshaw, A. A., D’Amico,
A. V., Richie, J. P., et al. (2002). Gene expression
correlates of clinical prostate cancer behavior. Cancer
cell, 1(2):203–209.
S
¨
onmez,
¨
O. S., DA
˘
GTEK
˙
IN, M., and Ensari, T. (2021).
Gene expression data classification using genetic
algorithm-basedfeature selection. Turkish Journal
of Electrical Engineering and Computer Sciences,
29(7):3165–3179.
Spira, A., Beane, J. E., Shah, V., Steiling, K., Liu, G.,
Schembri, F., Gilman, S., Dumas, Y.-M., Calner, P.,
Sebastiani, P., et al. (2007). Airway epithelial gene ex-
pression in the diagnostic evaluation of smokers with
suspect lung cancer. Nature medicine, 13(3):361–366.
Srinivas, M. and Patnaik, L. M. (1994). Genetic algorithms:
A survey. computer, 27(6):17–26.
Venkatesh, B. and Anuradha, J. (2019). A review of feature
selection and its methods. Cybernetics and informa-
tion technologies, 19(1):3–26.
Wang, S., Tang, J., and Liu, H. (2015). Embedded unsuper-
vised feature selection. In Proceedings of the AAAI
conference on artificial intelligence, volume 29.
Zebari, R., Abdulazeez, A., Zeebaree, D., Zebari, D., and
Saeed, J. (2020). A comprehensive review of di-
mensionality reduction techniques for feature selec-
tion and feature extraction. Journal of Applied Science
and Technology Trends, 1(1):56–70.
Zhao, W., Wang, L., and Zhang, Z. (2019). A novel atom
search optimization for dispersion coefficient estima-
tion in groundwater. Future Generation Computer
Systems, 91:601–610.
Improved Binary Elk Herd Optimizer with Fitness Balance Distance for Feature Selection Using Gene Expression Data
797
