
Preparing Ultrasound Imaging Data for Artificial Intelligence Tasks:
Anonymisation, Cropping, and Tagging
Dimitrios Pechlivanis
1 a
, Stylianos Didaskalou
2 b
, Eleni Kaldoudi
1,2 c
and George Drosatos
2 d
1
School of Medicine, Democritus University of Thrace, 68100 Alexandroupoli, Greece
2
Institute for Language and Speech Processing, Athena Research Center, 67100 Xanthi, Greece
Keywords:
Ultrasound Imaging, DICOM, Anonymisation, Cropping, Tagging, Artificial Intelligence (AI).
Abstract:
Ultrasound imaging is a widely used diagnostic method in various clinical contexts, requiring efficient and
accurate data preparation workflows for artificial intelligence (AI) tasks. Preparing ultrasound data presents
challenges such as ensuring data privacy, extracting diagnostically relevant regions, and associating contextual
metadata. This paper introduces a standalone application designed to streamline the preparation of ultrasound
DICOM files for AI applications across different medical use cases. The application facilitates three key pro-
cesses: (1) anonymisation, ensuring compliance with privacy standards by removing sensitive metadata; (2)
cropping, isolating relevant regions in images or video frames to enhance the utility for AI analysis; and (3)
tagging, enriching files with additional metadata such as anatomical position and imaging purpose. Built with
an intuitive interface and robust backend, the application optimises DICOM file processing for efficient inte-
gration into AI workflows. The effectiveness of the tool is evaluated using a dataset of Deep Vein Thrombosis
(DVT) ultrasound images, demonstrating significant improvements in data preparation efficiency. This work
establishes a generalizable framework for ultrasound imaging data preparation while offering specific insights
into DVT-focused AI workflows. Future work will focus on further automation and expanding support to ad-
ditional imaging modalities as well as evaluating the tool in a clinical setting.
1 INTRODUCTION
Machine learning (ML) has already achieved signif-
icant breakthroughs in medicine, especially in the
automated diagnosis in medical imaging, automated
management of administrative tasks, and exploration
of the structure of bio-molecules, particularly proteins
(Habehh and Gohel, 2021). A critical prerequisite
for the successful application of machine learning in
medical imaging is the preparation of high-quality, la-
belled datasets. This process, often referred to as ML
training data preparation, is particularly intricate due
to several factors: the need for strict anonymisation to
ensure patient privacy, the identification and extrac-
tion of diagnostically relevant image regions, and the
association of contextual metadata, such as anatom-
ical position and purpose. Ensuring patient privacy
through anonymisation is especially critical, as it sup-
ports both ethical considerations and compliance with
data protection regulations in medical research.
a
https://orcid.org/0009-0001-3583-772X
b
https://orcid.org/0000-0001-5932-9626
c
https://orcid.org/0000-0002-6054-4961
d
https://orcid.org/0000-0002-8130-5775
Medical imaging and signal data management is
almost exclusively based on the DICOM (Digital
Imaging and Communication On Medicine) image
and communication standard (ISO 12052, 2017). DI-
COM supports all medical imaging modalities but
also organises data into two key components: (1)
the metadata, which include patient-specific informa-
tion (e.g., name, date of birth, and hospital details),
as well as examination-specific data (e.g., modality
type, acquisition parameters), and (2) the imaging
data, which consist of pixel-based representations of
the medical image or video. In addition, some meta-
data fields may be “burnt-in” directly onto the imag-
ing data, as is common in ultrasound imaging, further
complicating the anonymisation process.
Traditional approaches to DICOM data prepara-
tion for ML training, including the removal or ob-
fuscation of sensitive metadata and annotations from
both the header and the image itself, are often man-
ual and time-intensive. These limitations hinder the
scalability of AI-based solutions in clinical practice,
underlining the importance of automated tools that
can handle both anonymisation and efficient dataset
preparation. This involves ensuring data privacy, stan-
dardising formats, isolating relevant image regions,
Pechlivanis, D., Didaskalou, S., Kaldoudi, E. and Drosatos, G.
Preparing Ultrasound Imaging Data for Artificial Intelligence Tasks: Anonymisation, Cropping, and Tagging.
DOI: 10.5220/0013379400003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 951-958
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
951

and enriching the data with metadata to meet the qual-
ity requirements for ML training.
Ultrasound imaging, as a widely used modality in
clinical practice, exemplifies these challenges due to
its operator-dependent variability and its application
across diverse diagnostic contexts. While these issues
are common across ultrasound imaging use cases,
they become particularly critical in specific condi-
tions such as deep vein thrombosis (DVT). DVT is
a serious medical condition characterised by the for-
mation of blood clots within deep veins, most com-
monly in the lower extremities (Waheed et al., 2024).
Ultrasound imaging is the primary diagnostic modal-
ity for DVT due to its non-invasive nature and real-
time imaging capabilities. However, the complex-
ity of analysing ultrasound data, coupled with vari-
ability in operator technique and patient anatomy,
poses challenges for consistent and accurate diagno-
sis (Baker et al., 2024). Artificial intelligence (AI)
offers a promising approach to addressing these chal-
lenges, automating tasks such as vessel segmenta-
tion and thrombus detection (Maiti and Arunachalam,
2022).
This paper introduces a tool, called
US-DICOMizer, to automate and streamline the
preparation of ultrasound DICOM files for AI-based
workflows. While the tool is designed for general
ultrasound imaging applications, it is evaluated using
a dataset of DVT ultrasound exams as a test case.
The application incorporates three key functionali-
ties: (1) anonymisation to remove sensitive patient
information while preserving essential metadata for
AI tasks, (2) cropping to extract relevant regions
from images or videos, and (3) tagging to annotate
files with critical metadata, such as anatomical
position, imaging purpose, and other contextual
information. These functionalities aim to address
the unique requirements of ultrasound imaging data
preparation while ensuring compliance with data
privacy regulations and clinical standards.
This work is part of the ThrombUS+ project, co-
funded by the European Union (EU) under Grant
Agreement No. 101137227 (Kaldoudi et al., 2024).
The ThrombUS+ initiative aims to develop an innova-
tive, operator-independent wearable diagnostic device
designed to facilitate the early detection and diagno-
sis of DVT using various diagnostic data modalities,
including ultrasound imaging. As part of this initia-
tive, the US-DICOMizer application addresses criti-
cal data preparation needs for AI-based workflows in
DVT and other ultrasound imaging applications.
The remainder of this paper is organised as fol-
lows: Section 2 provides an overview of related work
in DICOM data preparation and AI in ultrasound
imaging. Section 3 details the methodology and
design of the application, including its anonymisa-
tion, cropping, and tagging functionalities. Section 4
discusses the technical implementation details, high-
lighting the tools and frameworks employed. Section
5 presents results demonstrating the performance and
the user experience of the application. Finally, Sec-
tion 6 concludes the paper by summarising the contri-
butions of this work to AI-based medical imaging and
provides future directions.
2 RELATED WORK
The preparation of ultrasound imaging data for artifi-
cial intelligence (AI) tasks requires specialised tools
to address the unique challenges of this modality.
Anonymisation, cropping, and tagging are critical
steps to ensure patient privacy, enhance data utility,
and facilitate effective data sharing. Existing solu-
tions have focused on general medical imaging or
specific modalities, often neglecting the particular re-
quirements of ultrasound imaging for tasks like vessel
segmentation and thrombus detection. This section
reviews related work on anonymisation, cropping,
and tagging techniques in medical imaging, highlight-
ing their relevance and limitations in the context of
ultrasound.
Numerous anonymisation tools have been devel-
oped to safeguard patient privacy while ensuring com-
pliance with DICOM (Digital Imaging and Commu-
nications in Medicine) standards. Monteiro et al.
(2017) introduced a de-identification pipeline lever-
aging convolutional neural networks (CNNs) for char-
acter recognition, achieving an anonymisation rate of
89.2%. Similarly, Rodr
´
ıguez Gonz
´
alez et al. (2010)
developed an open-source toolkit for DICOM data
de-identification tailored to multicenter trials, offer-
ing customisation based on specific privacy needs.
However, these tools are primarily designed for neu-
roimaging and lacked features specific to ultrasound
workflows. Haselgrove et al. (2014b) presented a sys-
tem for anonymising DICOM data in neuroimaging
research, ensuring compliance with quality control
standards but leaving the unique needs of ultrasound
imaging unaddressed.
Cropping and tagging techniques play a cru-
cial role in isolating diagnostically relevant regions,
anonymising burned-in annotations text, and asso-
ciating metadata with medical images. Interactive
anonymisation tools, such as the Cornell Anonymi-
sation Toolkit (Xiao et al., 2009), allow users to man-
ually crop and anonymise datasets but do not offer
automation or optimisation for ultrasound imaging.
Tools like DICOM Anonymiser and dicom-anon4
support tagging within the framework of DICOM
standards (Haselgrove et al., 2014a), but their focus
on neuroimaging limits their relevance to ultrasound-
specific needs. More recently, image cropping
techniques based on grid-anchor formulations have
HEALTHINF 2025 - 18th International Conference on Health Informatics
952

emerged in general computer vision (e.g., CNN-based
methods), but these approaches are designed for aes-
thetic image processing rather than clinical applica-
tions (Zeng et al., 2022).
Despite these advancements, significant gaps re-
main in addressing the specific requirements of ul-
trasound imaging. Existing tools are often modality-
specific and fail to provide comprehensive solutions
that integrate anonymisation, cropping, and tagging
for ultrasound data. Furthermore, inconsistent stan-
dards across imaging communities pose challenges to
the adoption of unified workflows. This paper ad-
dresses these challenges by introducing a tool, as a
standalone application, tailored to the preparation of
ultrasound DICOM data for AI tasks. While the tool
is designed to be broadly applicable across ultrasound
imaging use cases, its functionality is evaluated using
deep vein thrombosis (DVT) examinations as a test
case to validate its performance and utility.
3 METHODOLOGY
The development of US-DICOMizer is driven by
the broader requirements of ultrasound imaging data
preparation for artificial intelligence (AI) tasks, while
incorporating insights from the ThrombUS+ project
(Kaldoudi et al., 2024). As part of this initiative,
Clinical Study A (Drougka, 2024), described in de-
tail in the data collection manual available at https:
//app.thrombus.eu/studies/a/, is used to validate its
functionality. The methodology combines insights
from established imaging protocols with tailored tool
development to streamline key data preparation steps,
such as anonymisation, cropping, and tagging. It ex-
cludes the labelling and annotation of the collected
images and videos, which will be performed as a sep-
arate and necessary step for AI training.
Requirements and Design Principles. Clinical
Study A outlines a structured process for ultrasound
data collection, emphasising accurate imaging of spe-
cific anatomical sites, tagging critical metadata, and
ensuring privacy compliance. This process inspired
the design of US-DICOMizer, following a user-
centred approach which automates data preparation
to reduce manual effort while preserving the integrity
and diagnostic value of the images and videos. The
tool adheres to the following principles:
1. Data Privacy Compliance: Adherence to
anonymisation standards, ensuring compatibility
with HIPAA and GDPR.
2. Standardisation and Consistency: Integration
of tagging functionalities aligned with DICOM
standards to support AI-based downstream tasks.
3. Workflow Efficiency: Minimising manual inter-
vention while enabling flexibility through interac-
tive features.
4. Automation and Scalability: Streamlining
repetitive tasks such as cropping and anonymisa-
tion to improve efficiency across large datasets.
5. Clinical Relevance: Supporting tagging based on
the anatomical positions and diagnostic context
detailed in the Clinical Study A protocol.
Data Collection Context. In Clinical Study A,
healthcare professionals collect ultrasound images
and videos following a structured protocol to assess
key venous sites in the lower extremities. The imag-
ing included:
• Compression Ultrasound: Capturing frames and
videos to evaluate vein compressibility at specific
sites (e.g., common femoral vein, popliteal vein).
• Color Doppler Ultrasound: Enhancing diagnostic
detail for specific anatomical regions.
• Optional Imaging: Capturing pathological sites
(e.g., thrombus visualisation) where necessary.
US-DICOMizer integrates these requirements by
providing a robust framework for importing, crop-
ping, tagging, anonymizing, and exporting DICOM
files, preserving the diagnostic and research value of
the data.
Workflow. The workflow followed for preparing DI-
COM files with US-DICOMizer is presented in Fig-
ure 1 and involves the following key steps:
1. File Loading and Validation: Users can load sin-
gle DICOM files, entire folders, or compressed
archives. The tool validates the files to ensure
compatibility and displays their metadata for re-
view. Multiframe DICOMs are previewed frame-
by-frame to facilitate inspection.
2. Cropping and Tagging:
• Cropping: Users define relevant regions of
interest either manually through an interac-
tive GUI or by applying preset configurations.
Multiframe cropping ensures uniformity across
videos.
• Tagging: Metadata, including anatomical po-
sition (e.g., proximal, distal), the side (left or
right), and the image purpose, are applied to
each file.
3. Anonymisation: Sensitive information is auto-
matically removed or replaced, with user-defined
configurations specifying which tags are to be
deleted. The tool generates unique identifiers for
studies, series, and instances to ensure compliance
with data privacy regulations.
4. Export: Prepared DICOM files are exported as a
ZIP archive, maintaining a standardised structure
and ensuring compatibility with AI workflows.
Preparing Ultrasound Imaging Data for Artificial Intelligence Tasks: Anonymisation, Cropping, and Tagging
953
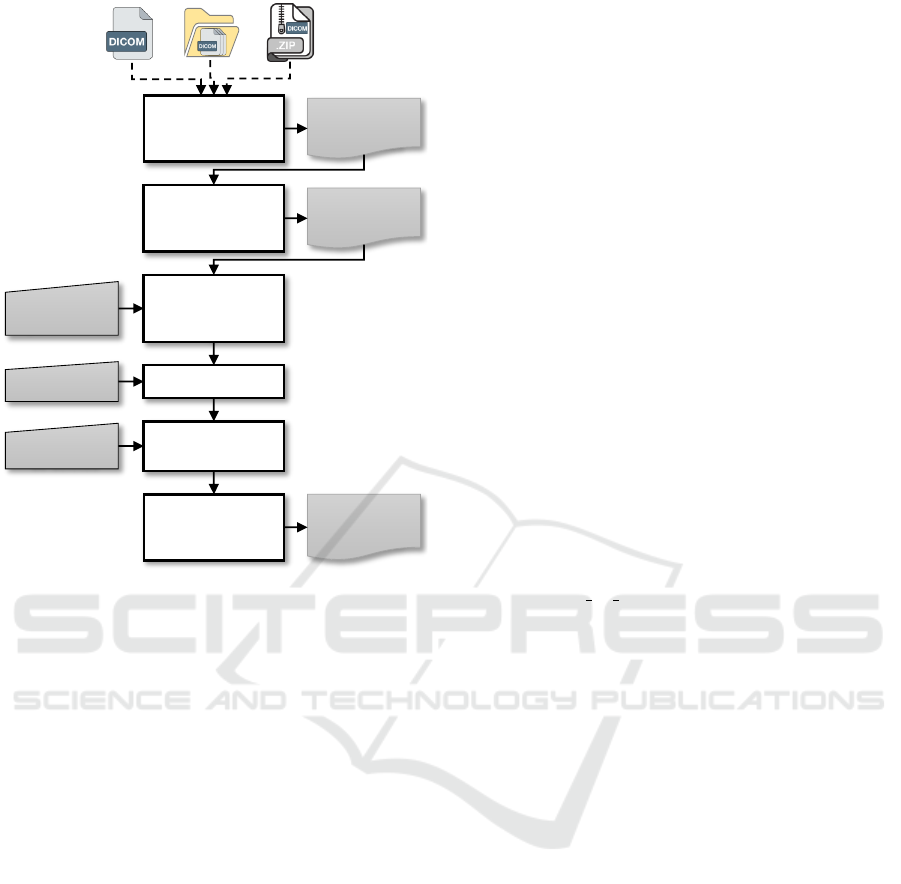
List of
available
DICOM files
Load and
validate DICOM
files
Cropping
parameters
Selection of
required
DICOM files
Final list of
DICOM files
Tags
Apply and
adjust cropping
region
Select tag
Apply
anonymisation
ZIP of
anonymized
DICOM files
Attributes to
be deleted
Export
anonymised
DICOM files
Figure 1: US-DICOMizer workflow.
By aligning the application workflow with this
protocol, US-DICOMizer bridges the gap between
clinical imaging practices and AI-driven research, en-
suring standardised, high-quality datasets.
4 IMPLEMENTATION DETAILS
The development of the US-DICOMizer focuses on
facilitating the preparation of ultrasound DICOM files
for artificial intelligence (AI) tasks through an in-
teractive and semi-automated workflow. The ap-
plication is open-source and available on GitHub
at https://github.com/thrombusplus/US-DICOMizer.
Below, we outline the technical details of its design
and implementation.
Development Environment and Frameworks. The
application is implemented in Python, leveraging the
following key libraries:
• Tkinter: For building a graphical user interface
(GUI) to facilitate intuitive user interactions.
• Pydicom: For reading, editing, and anonymising
DICOM files.
• SimpleITK: For image processing and visualisa-
tion of DICOM images.
• Pillow (PIL): For handling image formats and
performing operations like cropping and saving.
• NumPy: For efficient array manipulations of im-
age data.
• Matplotlib: For visualising images during pro-
cessing.
• Scikit-Image: For advanced image manipulation
and processing tasks.
Core Functionalities. Users can load individual DI-
COM files, entire folders, or ZIP archives containing
multiple DICOM files. Imported files are displayed in
a tree-view structure within the application, allow-
ing users to preview metadata and image content. For
multiframe DICOMs, a video slider facilitates frame-
by-frame visualisation.
Cropping regions are defined interactively through
the GUI or automatically based on pre-configured set-
tings in the settings.ini file. These cropping pa-
rameters can be applied uniformly to all frames of
multiframe DICOMs. Metadata tagging ensures the
association of critical information, including anatom-
ical position and image purpose, with each file. Crop
settings are also saved to the configuration file for
consistency across sessions.
Sensitive patient identifiers are removed or re-
placed during the anonymisation process. At-
tributes specified in the configuration file under the
[attributes to del] section are deleted from each
DICOM file. The application also generates new
unique identifiers (UIDs) for the study, series, and in-
stances to maintain data integrity while ensuring pri-
vacy compliance.
Processed DICOM files are exported as a com-
pressed ZIP archive. The naming conventions and
folder structure of the output are customisable via
the application settings, ensuring compatibility with
downstream workflows.
Error Handling and Logging. The application in-
corporates robust error handling to prevent workflow
interruptions. For instance:
• Validation of DICOM files ensures that non-
DICOM inputs are skipped during batch process-
ing.
• Invalid or missing cropping parameters result in
detailed error messages, guiding users to update
their configuration files.
• A dedicated logging system records application
activities and errors in a structured format for de-
bugging and audit purposes.
Customisability. User-defined settings, such as crop-
ping areas, compression levels, and attributes removal
lists, are stored in an INI configuration file. This file
allows for fine-grained control over the application’s
behaviour, enabling customisation to suit various use
cases and device configurations.
HEALTHINF 2025 - 18th International Conference on Health Informatics
954
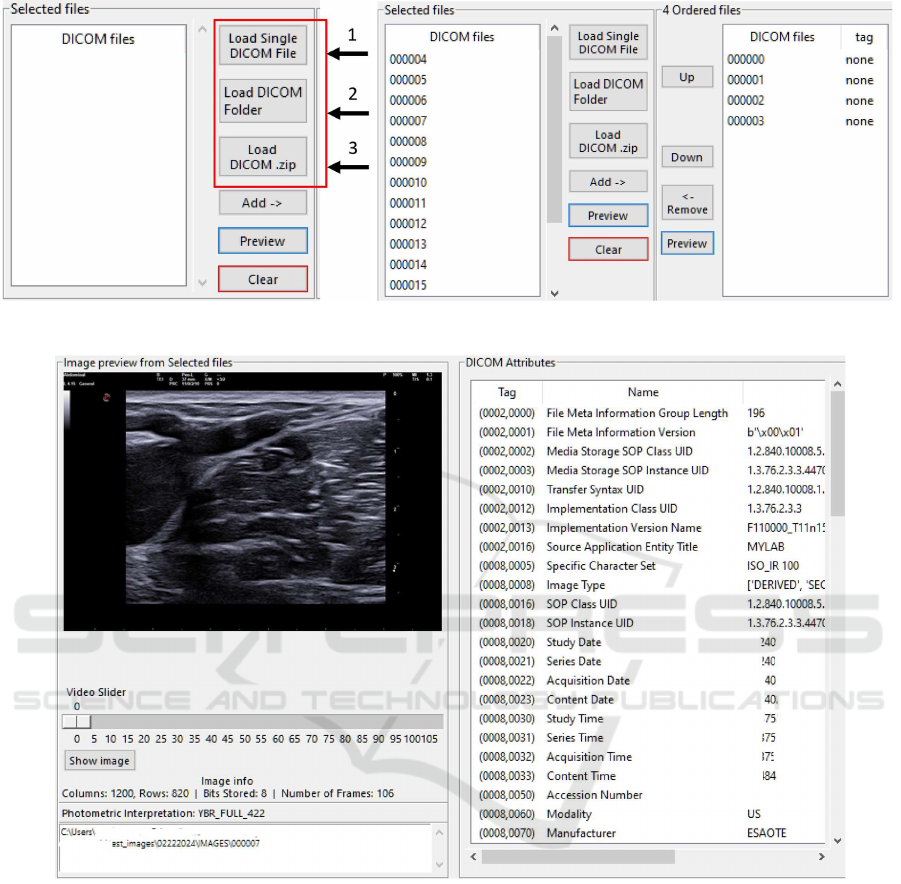
(a) Loading of DICOM files. (b) Selecting the appropriate DICOM files for anonymisation.
4
(c) Preview a DICOM file visually and as metadata.
Figure 2: Loading, selecting and previewing DICOM files.
Performance Optimisation. The application em-
ploys multithreading for tasks like previewing images
and processing large datasets, ensuring smooth opera-
tion without GUI freezes. Efficient handling of image
data through NumPy arrays and PIL ensures minimal
processing time.
5 RESULTS
The proposed US-DICOMizer application is evalu-
ated for its ability to streamline the preparation of ul-
trasound DICOM files for artificial intelligence (AI)
tasks through an intuitive sequence of steps: file load-
ing and preview, cropping and tagging, anonymisa-
tion, and export. Below, we describe the workflow
and outcomes achieved during testing.
Workflow Evaluation. Initially, users start the pro-
cess by importing DICOM files through multiple op-
tions, including selecting individual files, importing
entire folders, or uploading compressed archives (Fig-
ure 2.a). Imported files are displayed in the “Se-
lected File” list, allowing users to preview images and
inspect DICOM attributes, such as dimensions and
metadata (Figure 2.c). Multiframe DICOM files in-
clude an interactive video slider for frame-by-frame
Preparing Ultrasound Imaging Data for Artificial Intelligence Tasks: Anonymisation, Cropping, and Tagging
955
inspection. The interface provides users with the flex-
ibility to reorder, remove, or review files before fur-
ther processing (Figure 2.b).
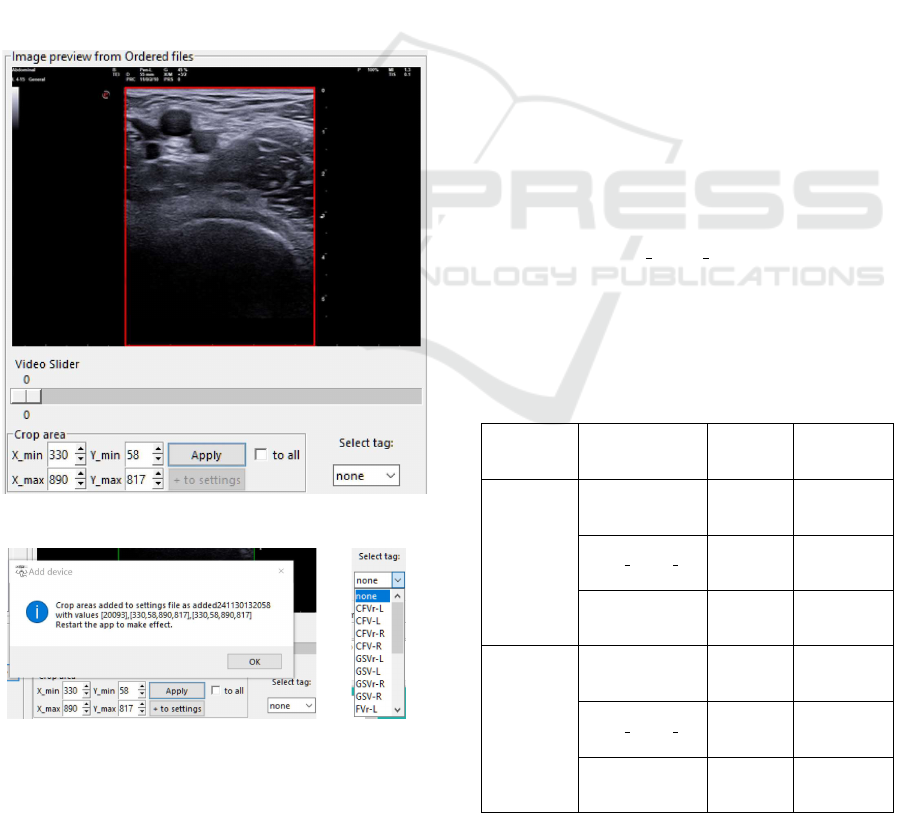
Once files are selected, users define cropping re-
gions to isolate diagnostically relevant portions of the
image or video frames. Predefined cropping settings
are automatically applied where applicable, with the
option for manual adjustment using an interactive pre-
view tool (Figure 3.a). Cropping parameters could be
applied uniformly across all frames or adjusted for
individual files, ensuring consistency and relevance
in data preparation. Additionally, cropping parame-
ters are saved to a configuration file, allowing users
to replicate consistent settings across multiple ses-
sions (Figure 3.b). Concurrently, users add tags to
each file to specify anatomical position (e.g., femoral
vein (FV) or popliteal vein (PV)), side (e.g., left or
right leg), and image purpose (e.g., diagnostic or non-
diagnostic quality) (Figure 3.c). These tags ensure
the association of critical metadata with the prepared
files, enhancing their utility for downstream AI tasks.
(a) Process of image cropping.
(b) Save cropping area in settings. (c) Select tag.
Figure 3: Cropping and tagging process.
With cropping and tagging completed, the
anonymisation process is initiated (Figure 4). The
application removes sensitive patient identifiers while
preserving essential metadata for AI processing. Files
with correctly defined cropping areas and tags are vi-
sually marked to indicate readiness for anonymisa-
tion. Validation checks prevented the anonymisation
of incomplete or improperly prepared files, maintain-
ing data integrity.
Anonymised DICOM files are exported as a com-
pressed ZIP archive for convenient sharing and stor-
age (Figure 5). Users specify the destination folder
for the exported archive, completing the preparation
process. This final step ensures that the files are ready
for integration into AI workflows with minimal man-
ual intervention.
Performance and User Experience. The applica-
tion demonstrates high efficiency and ease of use.
On average, ultrasound exams comprising 15-30 DI-
COM files (including single-frame and multiframe)
are fully prepared (including loading, cropping, tag-
ging, anonymisation, and export) within 3-6 minutes.
Exporting to a ZIP archive required less than 1–2 sec-
onds for exams of similar size.
The anonymisation process was evaluated for
its average execution times per frame across var-
ious photometric interpretations and cropping ar-
eas, as shown in Table 1. For single-frame ultra-
sound images with a resolution of 1200x800 pixels,
Monochrome2 photometric interpretation required
19–23 milliseconds, depending on the cropping area.
In contrast, the YBR FULL 422 and RGB interpre-
tations took longer, with times ranging from 25–30
milliseconds and 49–61 milliseconds, respectively.
For ultrasound multiframe DICOMs, the processing
Table 1: Average execution times (per frame) of anonymi-
sation process for various photometric interpretations and
cropping areas.
Media
Type
(resolution)
Photometric
Interpretation
Cropping
Area
(pixels)
AVG Time
per Frame
(msec)
Ultrasound
Image
(1200x800)
[100 runs]
Monochrome2
576x432 19
768×576 21
1024×768 23
YBR FULL 422
576x432 25
768×576 27
1024×768 30
RGB
576x432 49
768×576 53
1024×768 61
Ultrasound
Multi-frame
(1200x800)
[10 runs]
Monochrome2
576x432 102
768×576 105
1024×768 108
YBR FULL 422
576x432 117
768×576 119
1024×768 124
RGB
576x432 241
768×576 244
1024×768 250
HEALTHINF 2025 - 18th International Conference on Health Informatics
956
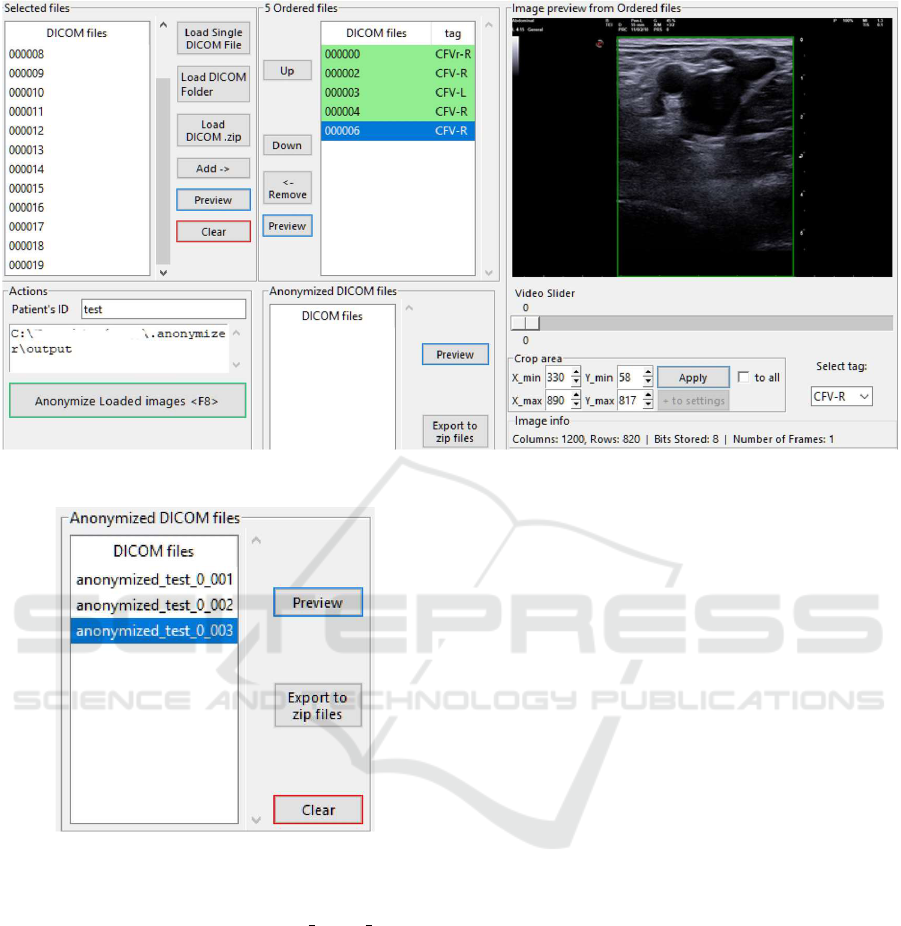
Figure 4: Start the anonymisation process after properly cropping and tagging the required DICOM files.
Figure 5: Export anonymised DICOM files as a ZIP file.
times increased as expected: Monochrome2 required
102–108 milliseconds per frame, YBR FULL 422 re-
quired 117–124 milliseconds, and RGB interpretation
ranged from 241–250 milliseconds per frame.
These results were produced using ultrasound ex-
ams from eight different ultrasound medical devices
manufactured by four vendors, and calculated on a
laptop computer running Windows 10, with an In-
tel Core i7-7700HQ CPU at 2.8 GHz and 16 GB of
RAM. While DVT datasets were used for testing, the
tool is designed for broader ultrasound imaging use
cases. Conclusively, this experiment highlights that
performance trade-offs are associated with both pho-
tometric complexity and cropping dimensions.
User feedback emphasises the application’s intu-
itive interface and streamlined workflow. Key fea-
tures, such as interactive previews before and after
anonymisation as well as cropping and tagging tools,
contribute to user satisfaction. Keyboard shortcuts
for common tasks further enhanced efficiency, mak-
ing the application a practical tool for medical imag-
ing professionals.
6 CONCLUSIONS AND FUTURE
WORK
The development and evaluation of US-DICOMizer
underscore its effectiveness in streamlining the prepa-
ration of ultrasound DICOM files for artificial intelli-
gence (AI) tasks. By automating key processes such
as anonymisation, cropping, and tagging, the appli-
cation addresses critical challenges in ultrasound data
preparation, including compliance with privacy stan-
dards, relevance of imaging regions, and metadata
contextualisation. The integration of these function-
alities into an intuitive, open-source platform facili-
tates scalable, standardised workflows for various ul-
trasound imaging AI applications. While the tool was
validated using deep vein thrombosis (DVT) datasets,
its design and functionality are adaptable to a wide
range of clinical use cases.
Preliminary results demonstrate that US-
DICOMizer significantly reduces manual effort
and enhances the consistency of prepared datasets,
enabling faster and more reliable AI model training.
The alignment of the tool’s design with the structured
protocol of Clinical Study A from the ThrombUS+
project ensures its clinical relevance and applicabil-
ity. Furthermore, its adaptability to various imaging
Preparing Ultrasound Imaging Data for Artificial Intelligence Tasks: Anonymisation, Cropping, and Tagging
957

workflows and compliance with industry standards
position it as a versatile tool for medical imaging
research.
Despite these achievements, several opportunities
for future development remain. First, expanding the
tool’s support to additional imaging modalities, such
as computed tomography (CT) or magnetic resonance
imaging (MRI), could broaden its utility. Second,
incorporating advanced AI algorithms directly into
the tool could enable automated cropping and tag-
ging based on learned patterns, further reducing the
need for manual input. Third, large-scale validation
of the tool in clinical settings is necessary to evaluate
its robustness and user satisfaction across diverse en-
vironments and datasets. Lastly, integrating real-time
feedback mechanisms and interoperability with eCRF
(electronic case report form) platforms could stream-
line data sharing, automate the collection process and
ensure compliance with clinical trial regulations.
In conclusion, US-DICOMizer represents a sig-
nificant step toward automating and standardising
ultrasound data preparation for AI-driven health-
care solutions. With planned enhancements and
broader adoption, it has the potential to accelerate ad-
vancements in diagnostic imaging and personalised
medicine, supporting a wide range of clinical and re-
search applications.
ACKNOWLEDGEMENTS
This work is co-funded by the European Union, un-
der the Horizon Europe Innovation Action Throm-
bUS+ (Grant Agreement No. 101137227). Views
and opinions expressed are however those of the au-
thors only and do not necessarily reflect those of
the European Union or HADEA as the granting au-
thority. Neither the European Union nor the grant-
ing authority HADEA can be held responsible for
them. Also, this work was carried out in the context
of the Inter-Institutional Master’s Program “Biomed-
ical Informatics” with the support of the School of
Medicine, Democritus University of Thrace and the
Athena Research Center in Greece.
REFERENCES
Baker, M., Anjum, F., and dela Cruz, J. (2024). Deep ve-
nous thrombosis ultrasound evaluation. In StatPearls.
StatPearls Publishing, Treasure Island (FL).
Drougka, I. (2024). D7.1: Clinical studies initiation pack-
age – Study A. ThrombUS+ Project, https://doi.org/
10.5281/zenodo.11371924.
Habehh, H. and Gohel, S. (2021). Machine learning in
healthcare. Current Genomics, 22(4):291–300.
Haselgrove, C., Poline, J.-B., and Kennedy, D. N. (2014a).
Comment on “A simple tool for neuroimaging data
sharing”. Frontiers in Neuroinformatics, 8.
Haselgrove, C., Poline, J.-B., and Kennedy, D. N. (2014b).
A simple tool for neuroimaging data sharing. Fron-
tiers in Neuroinformatics, 8.
ISO 12052 (2017). Health informatics – Digital imaging
and communication in medicine (DICOM) including
workflow and data management. Standard, Interna-
tional Organization for Standardization.
Kaldoudi, E., Marozas, V., Rytis, J., Pousset, N., Legros,
M., Kircher, M., Novikov, D., Sakalauskas, A., Mous-
takidis, P., Ayinde, B., Moltani, L. A., Balling, S.,
Vehkaoja, A., Oksala, N., Macas, A., Balciuniene,
N., Bigaki, M., Potoupnis, M., Papadopoulou, S.-
L., Grandone, E., Gautier, M., Bouda, S., Schloetel-
burg, C., Prinz, T., Dionisio, P., Anagnostopoulos, S.,
Drougka, I., Folkvord, F., Drosatos, G., Didaskalou,
S., and The ThrombUS+ Consortium (2024). To-
wards wearable continuous point-of-care monitoring
for deep vein thrombosis of the lower limb. In
Jarm, T.,
ˇ
Smerc, R., and Mahni
ˇ
c-Kalamiza, S., edi-
tors, 9th European Medical and Biological Engineer-
ing Conference, pages 326–335, Cham. Springer Na-
ture Switzerland.
Maiti, D. and Arunachalam, S. P. (2022). Non-invasive
diagnosis of deep vein thrombosis to expedite treat-
ment and prevent pulmonary embolism during gesta-
tion. volume 2022 Design of Medical Devices Con-
ference of Medical Devices, page V001T01A003.
Monteiro, E., Costa, C., and Oliveira, J. L. (2017). A
de-Identification pipeline for ultrasound medical im-
ages in DICOM format. Journal of Medical Systems,
41(5):89.
Rodr
´
ıguez Gonz
´
alez, D., Carpenter, T., Van Hemert, J. I.,
and Wardlaw, J. (2010). An open source toolkit for
medical imaging de-identification. European Radiol-
ogy, 20(8):1896–1904.
Waheed, S. M., Kudaravalli, P., and Hotwagner, D. T.
(2024). Deep vein thrombosis. In StatPearls. Stat-
Pearls Publishing, Treasure Island (FL).
Xiao, X., Wang, G., and Gehrke, J. (2009). Interactive
anonymization of sensitive data. In Proceedings of
the 2009 ACM SIGMOD International Conference on
Management of data, pages 1051–1054, Providence
Rhode Island USA. ACM.
Zeng, H., Li, L., Cao, Z., and Zhang, L. (2022). Grid anchor
based image cropping: A new benchmark and an ef-
ficient model. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 44(3):1304–1319.
HEALTHINF 2025 - 18th International Conference on Health Informatics
958
