
Comparative Analysis of CNNs and Vision Transformer Models for
Brain Tumor Detection
Safa Jraba
1a
, Mohamed Elleuch
2b
, Hela Ltifi
3c
and Monji Kherallah
4d
1
National School of Electronics and Telecommunications (ENETCom), University of Sfax, Tunisia
2
National School of Computer Science (ENSI), University of Manouba, Tunisia
3
Faculty of Sciences and Techniques of Sidi Bouzid, University of Kairouan, Tunisia
4
Faculty of Sciences, University of Sfax, Tunisia
Keywords: Brain Tumor, Deep Learning, Diagnosis, EfficientNet, ViT, Medical Imaging, Classification, SVM.
Abstract: Brain tumors are irregular cell mixtures existing within the brain or central spinal canal. They could be
cancerous or benign. The likelihood of the best possible prognosis and therapy increases with the speed and
accuracy of detection. This work provides a method for detecting brain tumors that combines the capabilities
of vision transformers and CNNs. In contrast to other studies that primarily relied on standalone CNN or ViT
architectures, our method uniquely integrates these models with a Support Vector Machine classifier for the
improvement of accuracy and robustness in medical image classification. While the ViT makes it possible to
combine CNN and ViT to improve the accuracy of medical imaging of the disease, the CNN extracts
hierarchical features. In-depth analyses of benchmark datasets pertaining to imaging modalities and clinical
perspectives were conducted. According to the experimental findings, ViT and EfficientNet identified tumors
with an accuracy of 98%, while the greatest reported accuracy of 98.3% was obtained when ViT was
combined with an SVM classifier. Our findings suggest that our method may improve brain tumor detection
methods.
1 INTRODUCTION
The abnormal growth of cells inside the brain or
Central Spinal Canal is the first indication of a brain
tumor. They fall into two categories: malignant-
invasive and benign-non-invasive. Benign tumors are
less aggressive because they do not contain cancer
cells, and they tend to have very well-defined
periphery patterns. Generally, they are amenable to
surgical interventions. Even benign tumors can
potentially wreak havoc on a patient's health by
compressing sensitive sites or interfering with the
circulation of the cerebrospinal fluid. Whereas
malignant tumors are composed of cancer cells, their
ability to invade local tumors and easily spread to
adjacent tissues renders them potentially deadly.
Radiation therapy, chemotherapy, and surgical
intervention may be provided as treatment options.
a
https://orcid.org/0009-0007-7818-5091
b
https://orcid.org/0000-0003-4702-7692
c
https://orcid.org/0000-0003-3953-1135
d
https://orcid.org/0000-0002-4549-1005
Extremely large computational efforts are often
required by this process-altering image encoders into
pixel sequences for their scrutiny. In response,
Parmar et al. (2018) considered self-attention in local
patches only around the query pixels while avoiding
the cost of global computations over the full image.
Meanwhile, for medical imaging, especially in
brain tumor detection and identification, there is no
little tribulation, heavily influencing treatment plans
and patient outcomes. Recent developments in deep
learning have fueled potential unlocks in the
evaluation of medical images, though tumor
diagnosis-based medical image analysis remains to
date, still an area earning its mark in practical
applications. CNNs have shown promise in feature
extraction and classification in many applications,
including medical imaging. In parallel, Vision
Transformers (ViTs) appeared and became popular
1432
Jraba, S., Elleuch, M., Ltifi, H. and Kherallah, M.
Comparative Analysis of CNNs and Vision Transformer Models for Brain Tumor Detection.
DOI: 10.5220/0013381900003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 3, pages 1432-1439
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

because of their capability to extract distant
associations from image data, which affords a unique
interpretation methodology.
Medical imaging experts continue to have serious
challenges in the identification and diagnosis of brain
tumors, which accordingly has a profound impact on
treatment and patient outcomes. Deep learning
techniques have come recently with a whole different
ball game in medical image processing, allowing
pathways towards accuracy and efficiency in tumor
diagnosis that have never been encountered. These
provide effective tools for feature extraction and
classification in different medical imaging fields
using CNNs. Concurrently, Vision Transformers
(ViTs) are mushrooming into popularity for excelling
at extracting distant associations from image data;
hence, offering a potentially new abstraction of image
interpretation.
Especially, this paper aims to do complete and
thorough and full measure-in-dependent study on the
CNN and Vision Transformers for cerebral tumor
detection in medical images. Such systems regain
their respective advantages by using each of the
architectures' strengths for a more resilient tumor
detection system and more accurate local-based
statistics. With the proposed method, we intend to
utilize all possible local and global information in the
medical images concurrently to deliver more detailed
and accurate tumor structure analyses.
In this work, we carry out comprehensive studies
on standard brain tumor picture datasets, comparing
our hybrid approach's efficacy to both standalone
Vision Trans-former models and traditional CNN-
based techniques.
Our results show the potential for CNN
(EfficientNet B0) and Vision Transformer fusion
technique to improve the diagnostic accuracy and
simplify the clinical decision-making process, as
evidence by the usefulness of both methods in brain
tumor detection. We speculated about the effects this
work could have on future medical imaging research
and presented suggestions for improving techniques
for brain tumor detection.
Contributions
This paper makes the following contributions:
- Provide a hybrid approach for brain tumor
detection that combines EfficientNet and
Vision Transformers (ViT) with SVM.
- A thorough comparison of CNN, solo ViT,
and our suggested hybrid model.
- Reach cutting-edge outcomes on MRI
datasets with an accuracy of 98.3%.
2 RELATED WORKS
The search for precise detection methodologies for
cerebral tumors has sparked significant research
efforts in recent years. Numerous approaches have
been investigated to address this vital need in the field
of health. Conventional diagnostic modalities such as
magnetic resonance imaging (MRI) and computed
tomodensitometry (CT scans) have historically been
the primary tools used to identify brain tumors. Their
effectiveness in early detection and precise
delineation of terrorist borders, however, remains a
challenge.
To identify and comprehend these cerebral
tumors, specialized ondelettes and sup-port systems
have been used in conjunction with MRI. The precise
and automated classification of brain IRM pictures
holds significant value in medical research and
interpretation. Uncontrolled cell division that results
in aberrant cell clusters inside or outside the brain is
the cause of brain tumors. These aberrant cell clusters
harm healthy cells and interfere with regular brain
function. The objective was to distinguish between
brain tissue that was unaffected by tumors and brain
tissue that had tumors, whether benign or malignant.
Gurbină et al. (2019)
CNN applied to MRI images has been shown to
be beneficial in many recent studies for the
classification of brain-related illnesses. Yuan et al.
(2018).
Researchers and medical professionals can locate
the area of the brain afflicted by a tumor by using
MRI, an imaging method that shows the anatomy and
structure of the human brain. Sakhthidasan et al.
(2021)
Abd-Ellah et al. (2019) have presented an
enhanced method for detecting brain tumors in order
to identify malignant tumors. Due to the low contrast
of mous tissues, lesion detection is a challenging task
that requires the use of adaptive clustering k-means to
obtain a better segmentation method in order to
improve prediction accuracy.
A thorough review of well-known deep learning
models that are applied to various types of brain
tumor investigation by Waqas et al. (2020).
Amarapur et al. (2019) discussed both traditional
automatic learning methods and deep learning
techniques for the validation of cerebral tumors. With
the aid of deep learning approaches, they were able to
identify, segment, and classify brain tumors
effectively using three different algorithms, leading to
improved performance.
A high-level System Cancer Diagnosis by
coalescing the four Level-I taxonomy components
Comparative Analysis of CNNs and Vision Transformer Models for Brain Tumor Detection
1433
"DIV" Data, Image Segmentation processing and
VIEW is proposed by Laukamp et al. (2019). Level-I
taxonomy DIV evaluation consists of acceptance
Rate and Completion Rate by DL convoluiton neural
networks.
Archana et al. (2023) performed a comparative
study of several optimizers used in convolutional
neural networks (CNNs) to detect brain tumors from
medical photos. The study examined how well the
Adam and SGD optimizers performed when applied
with the AlexNet and Le-Net CNN architectures. The
study's findings, which were based on a dataset of
1547 images, showed that the AlexNet architecture
and Adam optimizer outperformed LeNet and SGD to
obtain an average accuracy of 94.76%.
The use of deep neural networks and machine
learning methods for the early diagnosis of brain
tumors using MRI data was investigated by Wani et
al. (2023). A variety of CNN architectures, including
AlexNet, GoogleNet, VGG-19, a bespoke model, and
a collection of machine learning models, were used in
the study. Significant emphasis was made on data
gathering, preprocessing, and classification
procedures in order to address class imbalance and
data heterogeneity. The group of models that attained
the maximum accuracy of 90.625% suggests that
combining deep and conventional machine learning
techniques has bright futures.
Rajinikanth et al. (2022) applied various pooling
techniques with the pre-trained VGG16 and VGG19
convolutional neural networks (CNNs) to identify
glioma and glioblastoma brain tumors from magnetic
resonance imaging (MRI) images. Employing the
ADAM Optimizer, the work has examined the
classification performances of the SoftMax, Decision
Tree (DT), k-Nearest Neighbors (KNN), and Support
Vector Machine (SVM) classifiers on 2000 images
acquired from The Cancer Imaging Archive (TCIA).
The results showed that DT based on average-pooling
and VGG16 managed to gain maximum classification
accuracy, which was 96.08%.
Recent developments in Vision Transformers
(ViT) have shown that they may effectively extract
features from images by utilizing self-attention
techniques, outperforming conventional
convolutional neural networks. Zhang et al. (2O22)
In this section, we look closely at a few recently
published, more successful techniques (See Table 1).
Table 1: Research on the detection of brain tumors.
Authors Techni
q
ues Dataset Accurac
y
Archana et
al. (2023)
CNN,
AlexNet,
LeNet
1547
images of
Brain
Tumor
Dataset
94.76%.
Wani et al.
(2023)
AlexNet,
GoogleNet,
VGG-19,
Brain tumor
Dataset
MRI scans.
90.625%
Rajinikanth
et al. (2022)
VGG16
VGG19
SVM, KNN
MRI
images
96.08%
K.
Laukamp et
al. (2019)
Deep
learning
Model
Data View
(DIV)
95,08%
Our study produced impressive results that
outperformed previous methods. Through the
automation and optimization of the diagnostic
procedure, this research has a significant potential to
improve early brain tumor detection, which could
improve patient care and quality of life. Our findings
advance the state of the art in the categorization of
cerebral tumors and demonstrate the effectiveness of
transfer learning in the analysis of medical images.
3 METHODOLOGIES
This section describes the process of developing our
experiments to improve the accuracy of brain tumor
detection. The first step is to collect data. The second
stage involves data pre-processing, such as data
augmentation techniques. The last step is to predict
the outcome, where the ViT and EfficientNet B0
models are applied.
3.1 Gathering Data
The tomodensitometry (CT) and magnetic resonance
imaging (MRI) are two of the many biomedical
imaging techniques that are vital for the detection of
brain tumors. The MRI, which is especially well-
known for its high resolution and detailed
information, provides better viewpoints. Table 2
displays the MRI dataset from the well-known
Kaggle website, which had 3264 images categorized
into three groups.
Table 2 provides a clear overview of the training
and test data distribution across classes, now
including percentages to highlight data balance or
imbalance for glioma, meningioma, and pituitary
categories.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1434
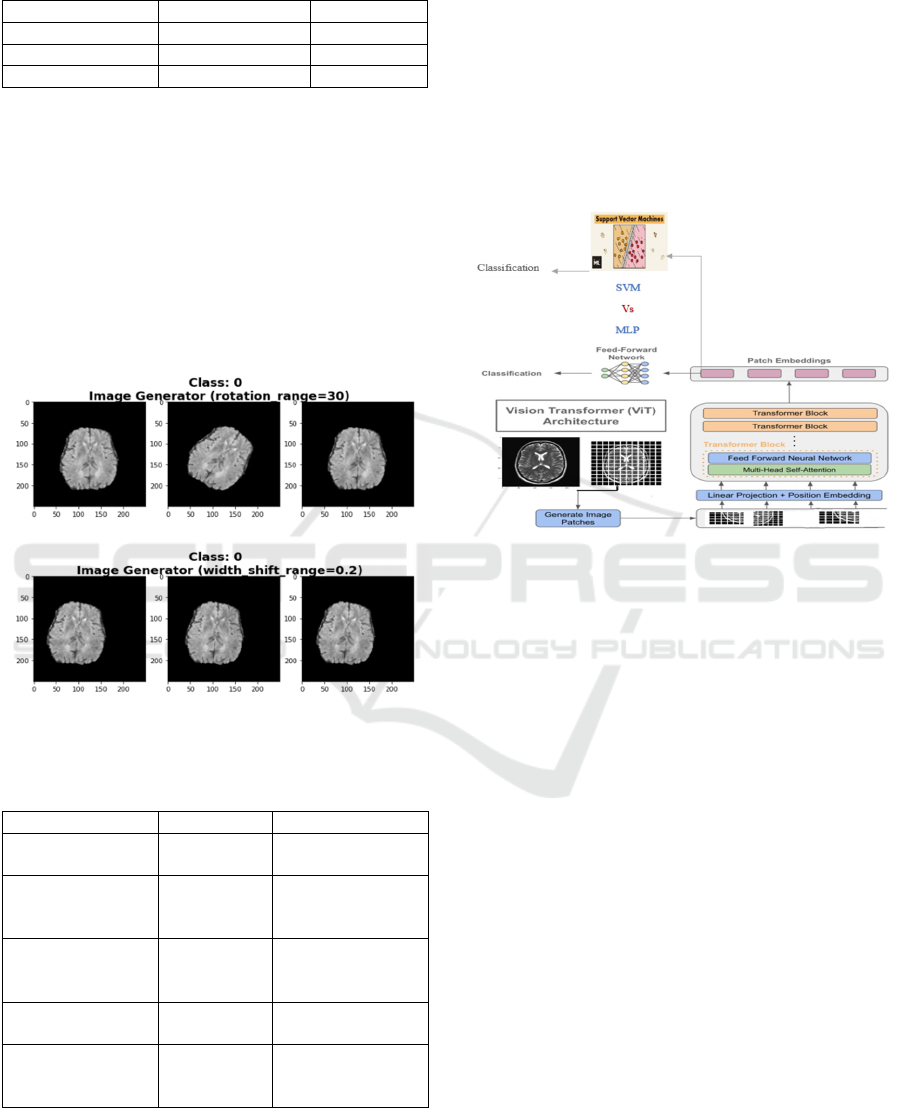
Table 2: Contents of a collection for brain MRI scans.
Data Label
Class Train set (%) Test set (%)
Glioma 40% 42%
Meningioma 35% 33%
Pituitar
y
25% 25%
3.2 Pre-Processing
Our brain tumor MRI dataset underwent a number of
pre-processing procedures to guarantee data
consistency and quality. In order to improve model
understanding and prediction accuracy, these steps
included resizing images to a standard dimension,
normalizing the data to improve model convergence,
and using augmentation techniques to diversify the
dataset and represent different tumor classes, such as
glioma, meningioma, pituitary (See Figure 1).
Figure 1: Using the MRI brain tumor data augmentation
technique.
Table 3: Augmentation techniques used.
Techni
q
ue Parameters Ob
j
ective
Rotation [-30°, +30°] Introduce angle
variations
Translation [-0.2, +0.2] Simulate
horizontal/vertical
shiftin
g
Scaling [0.8, 1.2] Represent
different tumor
sizes
Horizontal/vertic
al fli
p
Random Reduce
directional bias
Luminosity/
Contrast
[0.5, 1.5] Simulate different
lighting
conditions
The data augmentation methods used to increase
model generalization and resilience in brain tumor
detection tasks are shown in Table 3.
3.3 Proposed Architectures
3.3.1 Vision Transformer (ViT)
Vision Transformer model debuted at the 2021
International Conference on Learning Representations
(ICLR) in the paper "An Image is Worth 16x16 Words:
Transformers for Image Recognition at Scale." ViTs
adapt transformer topologies from natural language
processing (NLP) to convert input images into patches
resembling word tokens.
Figure 2: Vision Transformers (ViT) architecture.
Brain tumor images are converted into "patches"
for examination. The raw image is divided into small
sections prior to applying the "patches," which are
then processed by the ViT model to produce
insightful representations. This transformation
process, ViT can effectively identify and categorize
aspects of brain tumors as well as other relevant
components in the medical image.
For classification, the standard ViT employs a
Feed-Forward Network (FFN) (See Figure 2). In our
approach, we replaced the FFN classifier with a
Support Vector Machine (SVM) to leverage its proven
performance in the medical field. Yang et al. (2018)
The Support Vector Machine (SVM) classifier is
utilized for brain cancer detection due to its ability to
handle high-dimensional data and create a robust
decision boundary between different classes (See
Figure 3). Rajinikanth et al. (2022)
By finding the optimal hyperplane that maximizes
the margin be-tween tumor and non-tumor samples,
the SVM can accurately classify new, unseen images
as either containing a brain tumor or not. Its
effectiveness in medical applications, including
cancer detection, stems from its ability to minimize
classification errors and enhance predictive accuracy.
Garcia et al. (2020)
Comparative Analysis of CNNs and Vision Transformer Models for Brain Tumor Detection
1435

Figure 3: SVM approaches; (a) one-versus-all method, (b)
one-versus-one method. Tan et al. (2023).
3.3.2 EfficientNet B0
A convolutional neural network (CNN) with limited
processing and memory capacity, EfficientNet B0 is
intended for deep learning applications. Due to the
simultaneous optimization of the CNN models' depth,
width, and resolution, it has a lightweight but robust
architecture (See Figure 4).
Figure 4: Efficient Net B0 architecture.
With the release of EfficientNetV2, model
performance and training speed were enhanced,
making it especially appropriate for computationally
demanding jobs like medical imaging. Zhang et al.
(2023)
4 EXPERIMENTS AND RESULTS
This section describes the data collection process; the
images of cerebral tumors serve as the basis for the
data used in this work. Effective categorization
results are achieved when a broad and diverse data set
is used. We will describe and discuss the
experimental configurations in the following. The
obtained results are then presented and contrasted
with the suggested systems.
We investigate the potent capabilities of CNN and
Vision Transformers (ViT) architectures to solve the
challenging task of picture classification on the
cerebral tumor data game. Conventional
convolutional neural networks (CNNs) have been the
architecture of choice for image-related tasks, but
vision-related tasks (ViTs) introduce auto-attention
mechanisms inspired by the Transformer
architecture, which was originally designed for
natural language processing.
Furthermore, in order to actually carry out the
study, a phase of analysis and discussion of the
experiment's parameters is required (See Table 4).
Table 4: Parameters of EfficientNet B0 Model.
Total parameters 4,054,695
Trainable
p
arameters 4,012,672
Non-trainable
p
arameters 42,023
In this work, we combined a Support Vector
Machine (SVM) for classification with Vision
Transformers (ViT) as the foundation for feature
extraction. The following are specifics of the SVM
configuration that was used:
Kernel Function: Because radial basis
function (RBF) can handle nonlinear feature
spaces, that's the kernel we used.
Kernel Parameters for RBF:
Sigma: The value of sigma, also called
gamma, is fixed at 0.01. The influence of a
single training point is controlled by this
parameter.
Coefficient: A second parameter was set to
0.5. For some RBF kernel implementations, it
is commonly referred to as coef0. The impact
of the linear term in the polynomial and
sigmoid kernels is adjusted by this coefficient.
4.1 Dataset
An accurate dataset is essential for using CNNs to
classify brain tumors into Glioma, Meningioma, and
Pituitary categories. Our dataset, sourced from
medical facilities, includes three distinct labels for
tumor classification:
Glioma: classification for brain tumors with
gliomas as its defining feature.
Meningioma: The categorization of brain
tumors classified as meningiomas
Pituitary: Brain tumors associated with the
pituitary gland.
The gathered dataset is split into two sub-sets: the
first represents the 80% training portion and the
second, the 20% test portion. There are three classes
of images for each of the two image portions, and
each class corresponds to a distinct label for brain
tumor classification (See Figure 5).
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1436
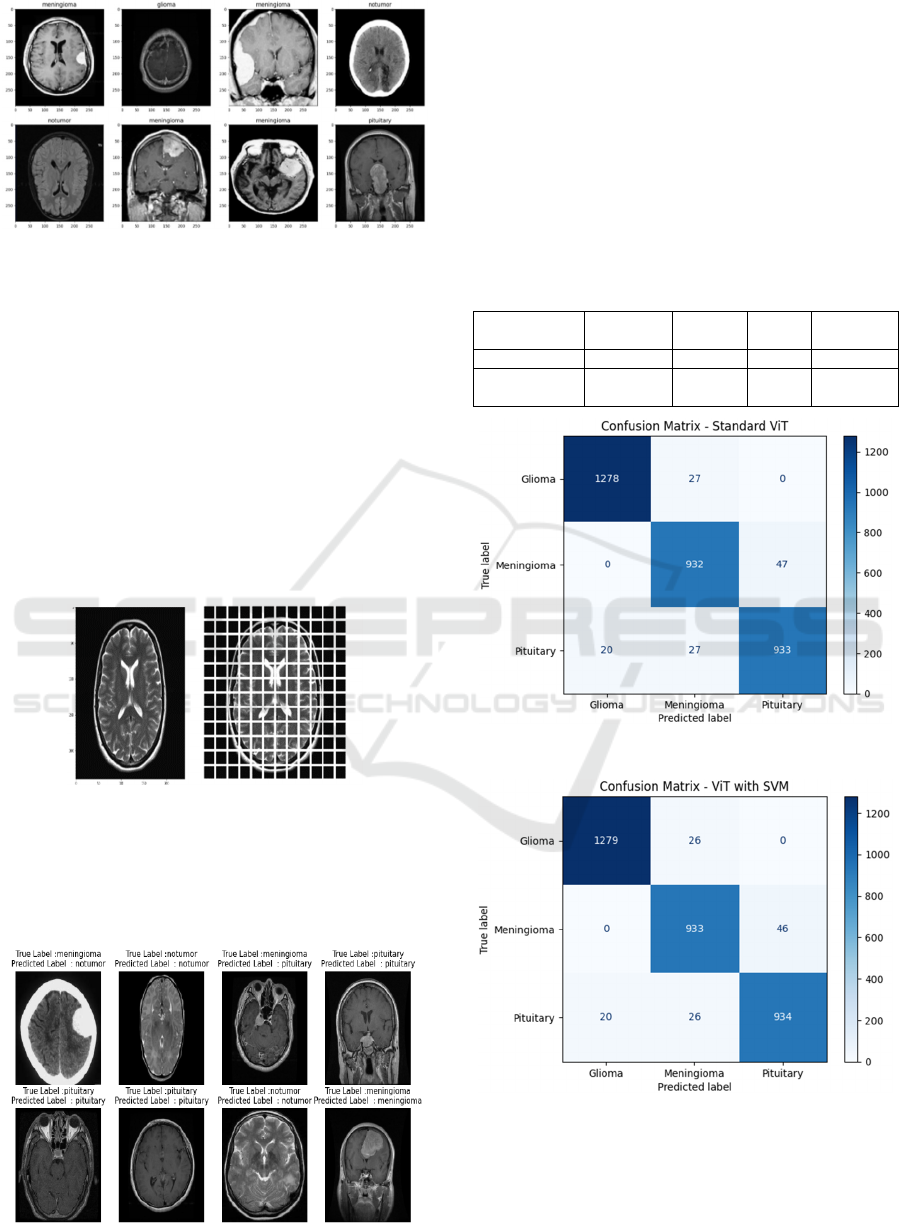
Figure 5: Samples of brain tumor labels.
4.2 Results and Discussion
One of the most widely used systems, the
Convolutional Neural Network (CNN) model, is used
to classify brain tumors into the Glioma, Meningioma,
and Pituitary classes. The performance of the CNN
convolutional neural network, the EfficientNet B0, and
the Vision Transformers (ViT) architectures are the
main subjects of the experimental investigation. The
following figures (6 and 7) provide a summary of our
suggested system's performance.
4.2.1 Vision Transformers ViT
Figure 6: Sample of Brain tumor image applying ViT.
Figure 6 illustrates a brain tumor MRI image
processed by the Vision Transformer (ViT), dividing
the image into grid-like patches for feature extraction
and classification.
Figure 7: Performance of ViT approach.
The standard ViT shows equable scores of 98% in
measures of precision, recall, F1 score, and accuracy,
demonstrating reliable classification. Using ViT and
SVM, we get an enhanced precision of 98.39% and an
accuracy of 98.3%; this means reduced false positives,
while recall drops slightly to 98.03%. The F1 slightly
increases to 98.2% signifying an overall improvement.
These metrics indicate that ViT can achieve very
good brain tumor detection by accurately
distinguishing tumor images from the rest of the
medical images. (See Table 5)
Table 5: Classification report of our proposed method (ViT).
Methods/
Metrics
Precision Recall
F1-
Score
Accuracy
Standard ViT 98% 98% 98% 98%
ViT with
SVM
98,39% 98,03% 98,2% 98,3%
Figure 8: Confusion matrices using ViT.
Figure 9: Confusion matrices using ViT combined with
SVM.
Figures 8 and 9 presents the confusion matrices
generated for the image brain tumor detection task
using ViT and ViT combined with SVM.
Comparative Analysis of CNNs and Vision Transformer Models for Brain Tumor Detection
1437
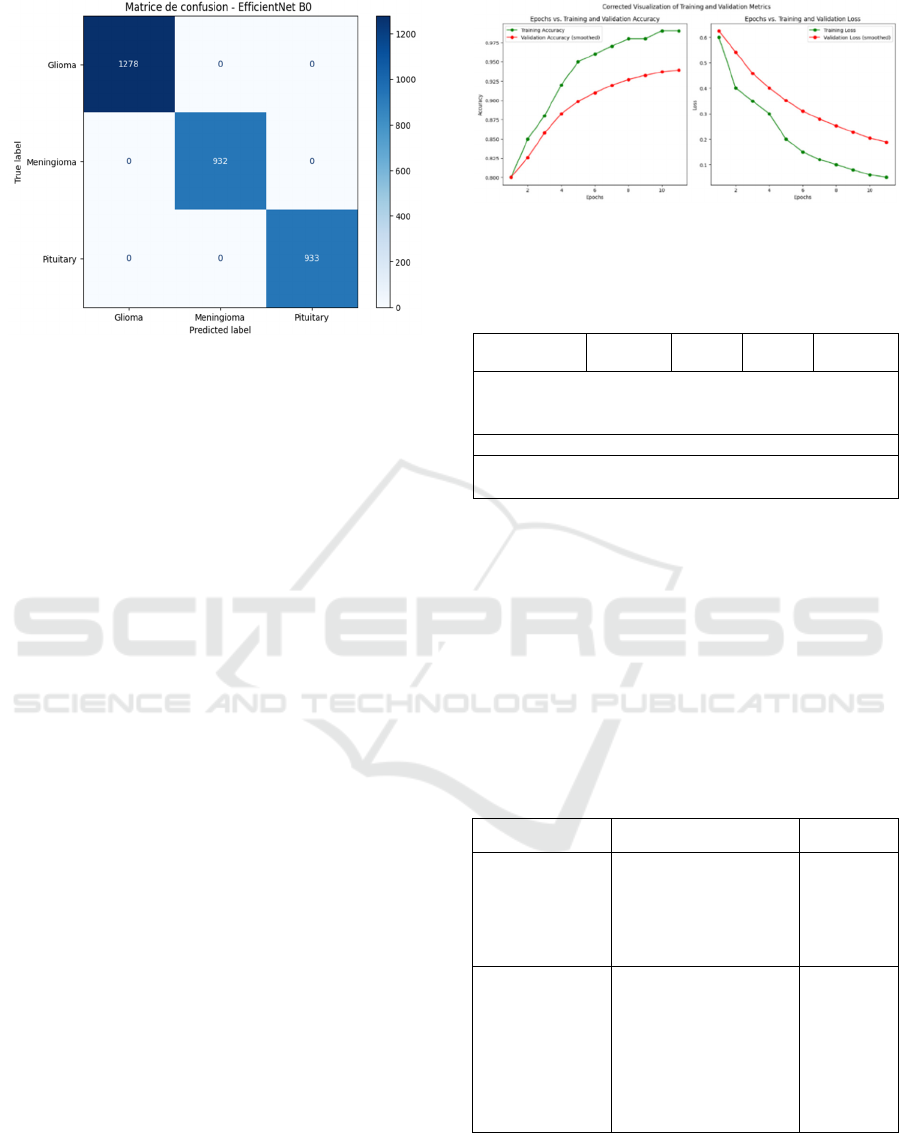
Figure 10: Confusion matrix of the proposed methods
(Efficient Net B0).
For the Standard ViT model, with perfect balance
between them. It predicts the majority of cases
correctly, and only makes minor errors, especially in
the Gliomas class because of its small number of false
negatives. On the whole, the model is performing
well, with only the slightest hit to certain classes.
All three metrics indicate extremely high
precision and recall (98%).
On the other hand, the ViT with SVM slightly
improves overall precision (98.39%) against the
Standard ViT according to incorrect predictions of a
small number of false negatives identified with the
Glioma class. In some classes, the performance on
Meningioma was a little compromised; however, this
model establishes a more balanced mechanism across
the classes, with an overall increase due to lesser
errors expected, thus proving this is an efficient and a
more reliable model to perform tumor detection.
4.2.2 Efficient Net B0
To evaluate a brain tumor detection model's efficacy,
its ability to classify gliomas, meningiomas, and
pituitary tumors is assessed. EfficientNet B0's
confusion matrix (Figure 9) shows true positives
(TP), true negatives (TN), false positives (FP), and
false negatives (FN). Metrics like precision, recall,
and specificity are computed to measure its
performance.
Plotting the metrics for the 98% accurate Efficient
Net B0 architecture would undoubtedly shed light on
the model's performance throughout evaluation and
training. Loss and additional measures like validation
accuracy, validation loss, and confusion matrix are
frequently presented alongside accuracy (See Figure
11).
Figure 11: Accuracy vs Epoch and Loss vs Epoch Graph
(Efficient Net B0).
Table 6: Classification report of our proposed methods
(Efficient Net B0).
Precision Recall
F1-
score
Support
Glioma
0.98 0.96 0.97 93
Meningioma
0.98 1.0 0.98 51
Pituitary
0.98 0.98 0.98 96
Accuracy
0.98 327
Macro avg
0.98 0.98 0.98 327
Weighted avg
0.98 0.98 0.98 327
The effectiveness of a brain tumor detection
model is assessed in this work. Accompanying the
support values for every class are f1-score, recall, and
precision metrics. The model's promise for brain
tumor diagnosis is demonstrated by its high precision,
recall, for all classes, as well as its overall accuracy
of 98% (see Table 6). The CNN and ViT hybrid
strategy make use of complementary strengths.
Likewise, SVMs continue to be reliable classifiers for
tasks involving medical imaging. He et al. (2022)
Table 7: Performance Comparison using Brain tumor
Dataset.
Authors Architecture Accuracy
Our Proposed
Vision Transformer
ViT
ViT with SVM
EfficientNet B0
98%
98.3%
98%
Wani et al.
(
2023
)
CNN, AlexNet, LeNet
94.76%
Rajinikanth et
al.
(
2022
)
AlexNet, GoogleNet,
VGG-19
90.62%
Zhang et al.
(
2022
)
VGG16, VGG19,
SVM, KNN
96.08%
Pang et al.
(
2023
)
Deep Learning Model 95.08%
Table 7 compares various brain tumor detection
methods. Our models (EfficientNet, ViT, and SVM)
achieved a maximum accuracy of 98.3%,
outperforming previous methods like VGG19,
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
1438

AlexNet, GoogleNet, and KNN. This demonstrates
the effectiveness of our lightweight and efficient
architectures, such as Net B0 and ViT, in achieving
promising results for early tumor diagnosis with high
accuracy.
4 CONCLUSIONS
Our investigation into EfficientNet and Vision
Transformer methods yielded a notable 98.3%
accuracy for brain tumor identification. By
combining Vision Transformer, SVM, and
EfficientNet, we developed a reliable MRI-based
detection system. Future work could explore Faster
R-CNN integration to improve the speed and
accuracy of tumor diagnosis, potentially enhancing
patient outcomes and advancing brain tumor
detection.
REFERENCES
N. Parmar, A. Vaswani, J. Uszkoreit, L. Kaiser, N. Shazeer,
A. Ku, and D. Tran. (2018).Image transformer,
International conference on machine learning. PMLR,
pp. 4055–4064
M. Gurbină, M. Lascu and D. Lascu. ( 2019 ).Tumor
Detection and Classification of MRI Brain Image using
Different Wavelet Transforms and Support Vector
Machines, 42nd International Conference on
Telecommunications and Signal Processing (TSP).
Yuan, Lin, et al.(2018).multi-center brain imaging
classification using a novel 3D CNN approach, IEEE
Access 6 , 49925-49934
K.Sakthidasan, A S Poyyamozhi, Shaik Siddiq Ali, and
Y.Jennifer. ( 2021).Automated Brain Tumor Detection
Model Using Modified Intrinsic Extrema Pattern based
Machine Learning Classifier Fourth International
Conference on Electrical, Computer and
Communication Technologies.
M. K. Abd-Ellah, A. I. Awad, A. A. Khalaf, and H. F.
Hamed. (2019). A review on brain tumor diagnosis
from MRI images: Practical implications, key
achievements, and lessons learned, Magnetic resonance
imaging, vol. 61, pp. 300-318.
M. Waqas Nadeem, Mohammed A. Al Ghamdi, Muzammil
Hussain, Muhammad Adnan Khan, Khalid Masood
Khan, Sultan H. Almotiri and Suhail Ashfaq
Butt.(2020). Brain Tumor Analysis Empowered with
Deep Learning: A Review, Taxonomy, and Future
Challenges Brain Sci.
B. Amarapur.(2019).Cognition-based MRI brain tumor
segmentation technique using modified level set
method, Cognition, Technology & Work, vol. 21, pp.
357-369.
K. R. Laukamp, F. Thiele, G. Shakirin, D. Zopfs, A.
Faymonville, M. Timmer, et al.(2019). Fully automated
detection and segmentation of meningiomas using deep
learning on routine multiparametric MRI, European
radiology, vol. 29, pp. 124-132.
B. Archana, M. Karthigha, and D. Suresh Lavanya. (2023).
Comparative Analysis of Optimisers Used in CNN for
Brain Tumor Detection, Institute of Electrical and
Electronics Engineers (IEEE).
S. Wani, S. Ahuja, and A. Kumar.(2023).Application of
Deep Neural Networks and Machine Learning
algorithms for diagnosis of Brain tumour, in
Proceedings of International Conference on
Computational Intelligence and Sustainable
Engineering Solution, Institute of Electrical and
Electronics Engineers Inc. pp. 106–111.
V. Rajinikanth, S. Kadry, R. Damasevicius, R. A. Sujitha,
G. Balaji, and M. A. Mohammed. (2022).Detection in
Brain MRI using Pre-trained Deep-Learning Scheme,
the 3rd International Conference on Intelligent
Computing, Instrumentation and Control Technologies:
Computational Intelligence for Smart Systems,
ICICICT
He, K., Zhang, X., Ren, S., & Sun, J. (2022). Synergizing
Convolutional and Transformer Models for Vision
Tasks. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 44(8), 4257-4268.
Yang, Z. R. (2018). Biological applications of support
vector machines. Briefings in bioinformatics, 5(4), 328-
338.
Jraba, S., Elleuch, M., Ltifi, H., & Kherallah, M. (2023).
Alzheimer Disease Classification Using Deep CNN
Methods Based on Transfer Learning and Data
Augmentation. International Journal of Computer
Information Systems and Industrial Management
Applications, 1-10..
Cervantes, J., Garcia-Lamont, F., Rodríguez-Mazahua, L.,
Lopez, A. (2020). A comprehensive survey on support
vector machine classification: Applications, challenges
and trends. Neurocomputing, 408, 189-2
Tan, M., Pang, R., & Le, Q. V. (2023). EfficientNetV2:
Smaller models and faster training. International
Conference on Machine Learning (ICML), 10023-
10033.
Comparative Analysis of CNNs and Vision Transformer Models for Brain Tumor Detection
1439
