
Histopathological Imaging Dataset for Oral Cancer Analysis: A Study
with a Data Leakage Warning
Marcelo Nogueira
1,3 a
and Elsa Ferreira Gomes
1,2 b
1
INESC TEC, Porto, Portugal
2
Instituto Superior de Engenharia do Porto, Porto, Portugal
3
Faculdade de Ci
ˆ
encias da Universidade do Porto, Departamento de Ci
ˆ
encia de Computares, Porto, Portugal
Keywords:
Oral Cancer, Histopathology, Deep Learning, CNN, Image Classification, Transfer Learning, Data
Augmentation, Data Leakage.
Abstract:
Oral squamous cell carcinoma is one of the most prevalent and lethal types of cancer, accounting for approx-
imately 95% of oral cancer cases. Early diagnosis increases patient survival rates and has traditionally been
performed through the analysis of histopathological images by healthcare professionals. Given the importance
of this topic, there is an extensive body of literature on it. However, during our bibliographic research, we
identified clear cases of data leakage related to contamination of test data due to the improper use of data
augmentation techniques. This impacts the published results and explains the high accuracy values reported
in some studies. In this paper, we evaluate several models, with a particular focus on EfficientNetBx architec-
tures combined with Transformer layers, which were trained using Transfer Learning and Data Augmentation
to enhance the model’s feature extraction and attention capabilities. The best result, obtained with the Effi-
cientNetB0, together with the Transformer layers, achieved an accuracy rate of 87.1% on the test set. To ensure
a fair comparison of results, we selected studies that we identified as not having committed data leakage.
1 INTRODUCTION
Oropharyngeal cancer ranks among the leading
causes of cancer-related deaths globally, particularly
affecting men. Its incidence varies widely based on
risk factors such as tobacco use, alcohol consumption,
poor oral hygiene, and limited access to healthcare.
In Europe, head and neck cancers account for approx-
imately 4% of all malignancies, with oral squamous
cell carcinoma (OSCC) being the most prevalent, oc-
curring in over 90% of patients diagnosed with head
and neck cancer (Vigneswaran and Williams, 2014).
The early detection and diagnosis of OSCC (Oral
Squamous Cell Carcinoma) significantly increases the
survival rate of patients and has traditionally been
carried out through the analysis of histopathologi-
cal images by health professionals. However, this
analysis is a demanding task for the medical team
(Chakraborty et al., 2019). Artificial Intelligence
techniques, specifically deep learning, help reduce di-
agnostic time and increase success rates (Fati et al.,
a
https://orcid.org/0000-0002-2776-900X
b
https://orcid.org/0000-0003-3610-8788
2022). Detecting OSCC through the classification of
histopathological images presents several challenges,
namely obtaining images with adequate quality. It is
also necessary to consider the heterogeneity of oral
carcinoma with a challenge factor, as it can be de-
tected in various shapes and sizes, as well as in dif-
ferent locations of the oral epithelium (Das et al.,
2023). Furthermore, developing deep learning mod-
els presents challenges, such as overcoming overfit-
ting and ensuring strong generalization, enabling the
model to perform well on data different from the
training set. Developing deep learning models capa-
ble of detecting oral cancer in histopathological im-
ages is expected to significantly aid the clinical com-
munity by enabling earlier diagnosis, improving pa-
tient survival rates, and facilitating faster and more
accurate diagnostic processes.
2 RELATED WORK
Most of the approaches found in the literature for
the detection of oral cancer in histopathological im-
Nogueira, M. and Gomes, E. F.
Histopathological Imaging Dataset for Oral Cancer Analysis: A Study with a Data Leakage Warning.
DOI: 10.5220/0013382100003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 811-818
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
811

ages include the use of a convolutional neural network
(CNN). Numerous studies have already proved the ef-
ficiency of computational histopathology applications
for automated tissue classification, segmentation, and
outcome prediction (Shavlokhova et al., 2021) (Soni
et al., 2024) (Nagarajan et al., 2023). In (Shavlokhova
et al., 2021), the authors study the application of a
CNN architecture (MobileNet) for automatized clas-
sification of oral squamous cell carcinoma from mi-
croscopic images. The proposed model achieved
an accuracy performance of 71.5% in the automated
classification of cancerous tissue. In (Nagarajan et al.,
2023), a deep learning framework was designed with
an intermediate layer between feature extraction lay-
ers and classification layers to classify histopatholog-
ical images as Normal and OSCC. The intermediate
layer is constructed using the proposed swarm intel-
ligence technique, called the Modified Gorilla Troops
Optimizer. To perform the feature extraction, the
use of three popular CNN architectures, namely, In-
ceptionV2, MobileNetV3, and EfficientNetB3. The
proposed methodology was evaluated in three pub-
lic dataset, and when they use the same dataset that
will be used in this work, the best result was 86% ac-
curacy. In (Soni et al., 2024), the authors tested 17
pre-trained deep learning models, to differentiate be-
nign and malign oral biopsy. For the different models
tested, they obtained accuracy results between 69.7%
and 86.7%, with the best result being obtained by the
EfficientNetB0 model. Our approach aims to lever-
age deep learning architectures based on CNNs with
transfer learning and Transformer layers. We will use
the Kaggle database (Kebede, 2021), ensuring proper
sampling of the available images to prevent data leak-
age. Data augmentation techniques will be applied to
balance classes, but only in the training set, ensuring
that the models are built robustly
1
.
2.1 Data Leakage Issues
During our bibliographic research, we identified clear
cases of different types of data leakage related to the
contamination of test data. The Histopathological
Imaging Database for Oral Cancer Analysis (Rahman
et al., 2020) consists of 1224 images (from 230 pa-
tients) divided into two sets with two different res-
olutions of the same images. The first set contains
89 histopathological images of normal oral epithe-
lium and 439 images of Oral Squamous Cell Carci-
noma (OSCC) at 100x magnification. The second set
consists of 201 images of normal oral epithelium and
495 histopathological images of OSCC at 400x mag-
1
https://www.kaggle.com/code/esterlita/efficientnetb0-
with-transformer
nification. In the literature, some studies report ap-
plying data augmentation to this dataset, increasing
its size from 1224 to 5192 images. This has led to
cases of data leakage, as observed in (Aiman, 2022),
(Ashraf, 2024), (Sharma, 2024) and (Rahman et al.,
2022), because they place data generated by data aug-
mentation in the validation and test sets, or because
synthetic data generated from the same original im-
age are placed in different sets (for example, in train-
ing and testing). These situations, where training im-
ages are inadvertently included in the test set, pro-
mote data leakage, and compromise confidence in the
reported results. We also identified cases of improper
dataset handling, such as applying data augmentation
to the entire dataset before the train/test split (Rahman
et al., 2022), or directly augmenting the test set, with
5000 samples subsequently reported (Albalawi et al.,
2024). Thus, we observed multiple cases of published
articles in which the results were positively biased due
to data leakage. However, in (Soni et al., 2024), a cor-
rect approach is evident: the train/test split was per-
formed before applying the data augmentation, multi-
ple models were tested using transfer learning and the
best result achieved was 86% accuracy with the Effi-
cientNetB0 model. Therefore, we will use this work
as a reference.
The dataset used in this work, available on Kag-
gle(Kebede, 2021), appears to have been derived from
the original dataset (Rahman et al., 2020) using data
augmentation. However, this information is not dis-
closed on the Kaggle platform.
3 METHODOLOGY
The methodology proposed for this study was to de-
velop deep learning architectures based on CNNs
with transfer learning and Transformer layers. The
methodology comprises two phases. In the first phase,
14 pre-trained CNN models were evaluated to detect
OSSC. In the second phase, the EfficientNetBx archi-
tecture models were explored, adding a Transformer
block to enhance attention capabilities, and evaluating
the impact this implementation has on model perfor-
mance.
3.1 Dataset
In recent years, two datasets with histopathological
images for oral cancer analysis have been made pub-
lic: Kaggle database (Kebede, 2021), and Histopatho-
logical database (Rahman et al., 2020). These two
data sets have served as the basis for the devel-
opment of OSCC identification algorithms through
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
812

Figure 1: Histopathological images of the class Normal
(the top figures) and the class OSCC (the bottom figures)
(Kebede, 2021).
histopathological images (Figure 1 shows examples
of each of the classes present in the dataset). In Table
1, we show the detailed datasets classes.
The Kaggle database appears to have been de-
rived from the Histopathological database, using data
augmentation. However, this information is not dis-
closed on the Kaggle platform. Thus, the kaggle
database contains 1224 images from the Histopatho-
logical database, plus 2204 images from the Normal
class and 1764 images from the OSCC class. These
images that were added to the Kaggle database were
generated using data augmentation techniques (rota-
tions, zooms, changes in luminosity, etc.), and are
contained in the database’s training set, with the pre-
fix ”aug ” before the name of the image. Therefore,
any research that uses the Kaggle database must adopt
the provided sample distribution for training, testing,
and validation, or exclude images generated through
data augmentation to prevent data leakage issues. In
this study, we used the Kaggle database. However,
since our goal was to modify the sample distribution
for training, testing, and validation, we had to exclude
images generated by data augmentation, leaving us
with the quantities from the original Histopatholog-
ical database (see the third column of Table 1).
With this amount of images, it was decided to cre-
ate balanced test and validation sets, as stated in Table
2. For the test set, 20% of the samples of the minority
class (0.2*290) and the same amount from the ma-
jority class were chosen. For the validation set, 10%
of the samples of the minority class (0.1*290) and
Table 1: Class distribution of the datasets.
Class Kaggle database
(Kebede, 2021)
Histopathological
database (Rah-
man et al., 2020)
Normal 2494 290
OSCC 2698 934
Total 5192 1224
the same amount from the majority class were cho-
sen. The remaining samples constitute the training
set. As at the moment the test and validation sets are
balanced, but the training set is not, it was decided
to balance the classes of the training set, generating
images of the OSCC class, through the data augmen-
tation technique (horizontal flips, vertical flips, and
zooms). Thus, 644 images from the OSCC class were
created to include in the training set, so that it was
also balanced.
Table 2: Distribution of images per subset.
Set Normal OSCC Total
Train 847 203 1050
Validation 29 29 58
Test 58 58 116
3.2 Technologies
The development of the deep learning model to clas-
sify histopathological images involved the use of ad-
vanced technologies that facilitate the construction,
training, and evaluation of neural network models.
This section details the tools and execution environ-
ment used, as well as their advantages and impact on
the project’s development. We used the Keras API
from Tensorflow and the Kaggle Notebook with a
GPU Tesla P100. The use of a Tesla P100 GPU is
of significant importance given the complexity of the
models that were tested. Several hyperparameters and
configurations were tested, with the aim of optimiz-
ing and making the models more robust. The Kag-
gle Notebook workflow, which uses the Keras API,
was designed for quick experimentation and iteration.
Data was processed directly in the environment, with
real-time visualizations of model training through ac-
curacy and loss graphs. Keras’s checkpointing and
callback features, combined with the GPU’s power,
enabled efficient model development.
3.3 Model Architecture
For the first phase, 14 pre-trained CNN models were
evaluated to detect OSCC, using the same dataset for
Histopathological Imaging Dataset for Oral Cancer Analysis: A Study with a Data Leakage Warning
813

each model. All the models used were pre-trained
on the ImageNet dataset. The pre-trained layers were
fine-tuned to capture relevant visual features, freezing
the lower layers while adjusting the top layers. The
14 models tested in this phase were: AlexNet (Alom
et al., 2018), Xception (Chollet, 2017), VGG16 and
VGG19 (Simonyan and Zisserman, 2015), ResNet50
and ResNet101 (He et al., 2016), DenseNet121,
DenseNet169 and DenseNet201 (Huang et al., 2017),
InceptionV3 (Szegedy et al., 2015), EfficientNetB0,
EfficientNetB1, EfficientNetB2 and EfficientNetB3
(Koonce, 2021).
In the second phase, the EfficientNetB0, Efficient-
NetB1, EfficientNetB2 and EfficientNetB3 architec-
ture models were explored, together with a Trans-
former block to enhance attention capabilities, allow-
ing the model to focus on different regions of the im-
age and capture global relationships between its parts.
Custom dense layers, along with L2 regularization
and Dropout, were included to prevent overfitting and
refine the features extracted by the convolutional and
attention layers. The architectural scheme of the pro-
posed model for the second phase is shown in Figure
2.
During image pre-processing, data augmentation
techniques were applied to the training set to increase
diversity (Torres et al., 2022) and improve the model’s
generalization ability (Figure 2). In particular, Hori-
zontal mirroring and vertical mirroring were applied,
reflecting the fact that important histopathological
features can appear in any orientation of the image.
This is a simple, yet effective transformation, partic-
ularly in the context of medical diagnosis, where the
orientation of features can vary without losing the se-
mantic content relevant to classification (Zeiser et al.,
2020).
In Table 3 we present the layers and hyperparam-
eters of the model and in Table 4 we present the com-
pilation and training settings of the proposed model.
In this approach, transfer learning was imple-
mented using the pre-trained EfficientNetB0 model,
whose architecture is represented in Figure 3, leverag-
ing the knowledge previously acquired from the Im-
ageNet dataset to improve the model’s accuracy and
efficiency in the task of classifying histopathological
images. This measure significantly reduced training
time and improved accuracy using feature extraction
from the large ImageNet dataset.
The Transformer block was introduced into the ar-
chitecture to complement the convolutional layers and
allow the model to learn attention over image fea-
tures. This block was inserted right after the output
of EfficientNetB0 and before the custom dense lay-
ers. This allowed the convolutional features learned
Table 3: Layers and hyperparameters of the model.
Layer Type Hyperparameters/
Description
Input Input Dimension:
(224,224,3) (RGB)
EfficientNetB0
(ImageNet)
Convolutional Only last 20 layers
unfrozen
for fine-tuning
Batch Normal-
ization
Normalization Epsilon=0.001
Momentum=0.99
Dense Dense 1024 neurons
Activation: ReLU
Regularization
L2=0.01
Dropout Dropout Rate=0.5
Dense Dense 512 neurons
Activation: ReLU
Regularization
L2=0.01
Dropout Dropout Rate=0.5
Dense Dense (output) 2 neurons
Activation: Soft-
max
Table 4: Compilation and training settings of the proposed
model.
Type Configuration / Description
Compilation Optimizer: Adamax
Learning Rate=0.001
Loss Function: Categorical Crossen-
tropy
Metrics: Accuracy
Train Batch Size=128
Epochs=50
Callbacks EarlyStopping: Monitoring validation
loss;
Patience=10
ReduceLROnPlateau: Monitoring val-
idation loss;
Factor=0.2;
Patience=2;
Minimum Learning Rate=1e-6
by EfficientNetB0 to be processed by the attention
layers, improving the model’s ability to capture global
relationships in the image before passing to the dense
layers. A Multi-Head Attention component was im-
plemented, enabling the model to focus on different
parts of the image simultaneously, allowing various
relationships between different regions of the image
to be modeled. The attention function considers dif-
ferent heads, or perspectives, of the image, learning
multiple representations at the same time. After the
attention layer, a feed-forward network was applied
to each position independently, allowing for the non-
linear transformation of the extracted features. The
feed-forward network was designed with dense lay-
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
814
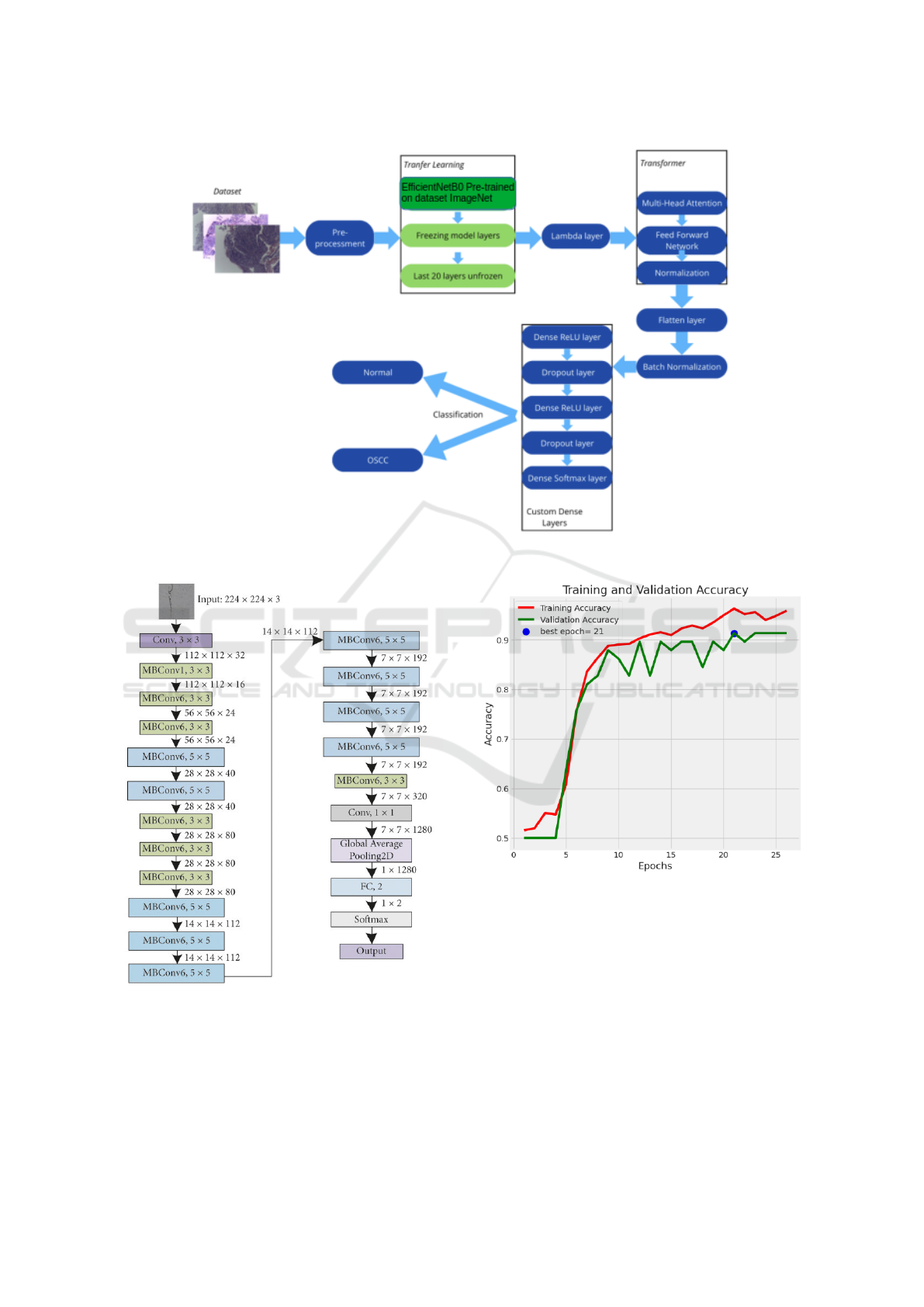
Figure 2: Model architecture.
Figure 3: EfficientNetB0 architecture model.
ers that helped refine the features after applying the
attention layer. To stabilize training and improve con-
vergence, layer normalization was applied after com-
bining attention and feed-forward components.
Figure 4: Accuracy of our model.
4 RESULTS AND DISCUSSION
As detailed before, the dataset has been split into
training, validation and test sets. Since the sets cre-
ated are class balanced, we can use accuracy for eval-
uating the model. In Table 5 we present the results
obtained for the 14 models used in the first phase,
and for the four models of the seconf phase, in which
the EfficientNetBX architecture was tested together
with a Transformer block. Our best result was ob-
Histopathological Imaging Dataset for Oral Cancer Analysis: A Study with a Data Leakage Warning
815
Table 5: Results of the several models used in test set.
Model Accuracy Sensitivity Specificity Precision Recall F1 Score
AlexNet 0.775 0.759 0.793 0.786 0.759 0.772
Xception 0.750 0.793 0.707 0.730 0.793 0.760
VGG16 0.741 0.741 0.741 0.741 0.741 0.741
VGG19 0.785 0.759 0,810 0.800 0.759 0.779
ResNet50 0.750 0.793 0.707 0.730 0.793 0.760
ResNet101 0.716 0.897 0.535 0.658 0.897 0.759
DenseNet121 0.759 0.776 0.741 0.750 0.776 0.763
DenseNet169 0.785 0.879 0.690 0.739 0.879 0.803
DenseNet201 0.785 0.862 0.707 0.746 0.862 0.800
InceptionV3 0.647 0.828 0.466 0.608 0.828 0.701
EfficientNetB0 0.793 0.672 0.914 0.886 0.672 0.765
EfficientNetB0 (Transformer) 0.871 0.759 0.983 0.978 0.759 0.854
EfficientNetB1 0.802 0.828 0.776 0.787 0.828 0.807
EfficientNetB1 (Transformer) 0.819 0.879 0.759 0.785 0.879 0.829
EfficientNetB2 0.776 0.897 0.655 0.722 0.897 0.800
EfficientNetB2 (Transformer) 0.819 0.776 0.862 0.849 0.776 0.811
EfficientNetB3 0.836 0.914 0.759 0.791 0.914 0.848
EfficientNetB3 (Transformer) 0.853 0.828 0.880 0.873 0.828 0.850
Table 6: Confusion Matrix of the EfficientNetB0 with
Transformer block.
Predicted:
Normal
Predicted:
OSCC
Actual:
Normal
57 1
Actual:
OSCC
14 44
tained with the EfficientNetB0 model, together with
the Transformer block, with an accuracy of 87,1% on
test set, with the results of the EfficientNetB3 model,
also with the Transformer block being very close to
this (accuracy of 85,3%). Analyzing the results ob-
tained, it can be seen that the models that present
the best performance are the models with the Ef-
ficientNetBx architecture. It can also be observed
that the inclusion of the Transformer block consis-
tently improves the model’s performance compared
to the models without it. The average accuracy of the
14 models that do not use the Transformer block is
76.4%, while the average accuracy of the models that
use the Transformer block is 84.1%. If we compare
only the four models of the EfficientNetBx architec-
ture, without the Transformer block, the average ac-
curacy of the models is 80.2%. The integration of the
Transformer block with the EfficientNetBx architec-
ture led to an average accuracy improvement of ap-
proximately 4%, demonstrating a positive impact on
the performance of the models.
Table 6 shows the confusion matrix of the Effi-
cientNetB0 model with the Transformer block, which
obtained the best result of all the models tested. In
Figure 4 we can see the evolution of our model’s
performance over the training and validation epochs,
which does not show signs of overfitting, as earlystop-
ping methodologies were used to monitor the model
training, evaluating the evolution of the model’s accu-
racy and loss in the validation set.
5 CONCLUSIONS AND FUTURE
WORK
The goal of our work was to contribute to the detec-
tion of oral cancer, specifically oral squamous cell
carcinoma (OSCC) using deep learning techniques.
We develop deep learning architectures based on
CNN’s with transfer learning and Transformer layers,
with special focus to the EfficientNetBx models. The
best result was obtained by the EfficientNetB0 model
together with the Transformer block, with an accu-
racy on the test set of 87.1%. The inclusion of the
Transformer block significantly improved the models’
accuracy, with an average increase of approximately
4% compared to the same models without the Trans-
former block.
We identified several studies in the literature that
use the same database as this work and present models
with excellent performance but are affected by multi-
ple types of data leakage. In this work, multiple types
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
816

of data leakage were identified in those studies, and as
a result, they were not considered for comparison of
results. However, we would like to highlight this issue
as a caution for future studies using these datasets.
For future work, we plan to adapt the model for
detecting oral cancer subtypes and incorporate image
segmentation techniques, which could enable more
precise identification of cancer-affected areas, thereby
complementing the clinical diagnosis process. The in-
tegration of this type of model into a clinical decision
support system is also a promising direction, with the
potential to improve the speed and accuracy of diag-
noses in hospital environments.
ACKNOWLEDGEMENTS
This work is financed by National Funds through
the Portuguese funding agency, FCT- Fundac¸
˜
ao
para a Ci
ˆ
encia e a Tecnologia, within project
LA/P/0063/2020. DOI:10.54499/LA/P/0063/2020.
REFERENCES
Aiman, E. (2022). Oral cancer, effcientnet classification,
98.7Accessed: December 14, 2024.
Albalawi, E., Thakur, A., Ramakrishna, M. T., Bhatia Khan,
S., SankaraNarayanan, S., Almarri, B., and Hadi, T. H.
(2024). Oral squamous cell carcinoma detection using
efficientnet on histopathological images. Frontiers in
Medicine, 10:3833.
Alom, M. Z., Taha, T. M., Yakopcic, C., Westberg, S.,
Sidike, P., Nasrin, M. S., Essen, B. C. V., Awwal, A.
A. S., and Asari, V. K. (2018). The history began from
alexnet: A comprehensive survey on deep learning ap-
proaches. ArXiv, abs/1803.01164.
Ashraf, K. (2024). Histopathologic oral cancer detection.
Accessed: December 14, 2024.
Chakraborty, D., Natarajan, C., and Mukherjee, A. (2019).
Chapter six - advances in oral cancer detection. vol-
ume 91 of Advances in Clinical Chemistry, pages
181–200. Elsevier.
Chollet, F. (2017). Xception: Deep Learning with Depth-
wise Separable Convolutions . In 2017 IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR), pages 1800–1807, Los Alamitos, CA, USA.
IEEE Computer Society.
Das, M., Dash, R., and Mishra, S. K. (2023). Auto-
matic detection of oral squamous cell carcinoma from
histopathological images of oral mucosa using deep
convolutional neural network. International Jour-
nal of Environmental Research and Public Health,
20(3):2131.
Fati, S. M., Senan, E. M., and Javed, Y. (2022). Early
diagnosis of oral squamous cell carcinoma based on
histopathological images using deep and hybrid learn-
ing approaches. Diagnostics, 12(8).
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In 2016 IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR), pages 770–778.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger,
K. Q. (2017). Densely Connected Convolutional Net-
works . In 2017 IEEE Conference on Computer Vision
and Pattern Recognition (CVPR), pages 2261–2269,
Los Alamitos, CA, USA. IEEE Computer Society.
Kebede, A. F. (2021). Histopathologic oral cancer detection
using cnns. Accessed: Jannuary 6, 2024.
Koonce, B. (2021). EfficientNet, pages 109–123. Apress,
Berkeley, CA.
Nagarajan, B., Chakravarthy, S., Venkatesan, V. K., Ra-
makrishna, M. T., Khan, S. B., Basheer, S., and Al-
balawi, E. (2023). A deep learning framework with an
intermediate layer using the swarm intelligence opti-
mizer for diagnosing oral squamous cell carcinoma.
Diagnostics, 13(22).
Rahman, A., Alqahtani, A., Aldhafferi, N., Nasir, M. U.,
Khan, M. F., Khan, M. A., and Mosavi, A. (2022).
Histopathologic oral cancer prediction using oral
squamous cell carcinoma biopsy empowered with
transfer learning. Sensors, 22(10).
Rahman, T. Y., Mahanta, L. B., Das, A. K., and Sarma,
J. D. (2020). Histopathological imaging database for
oral cancer analysis. Data in Brief, 29:105114.
Sharma, S. K. D. (2024). Oral squamous cell detection us-
ing deep learning.
Shavlokhova, V., Sandhu, S., Flechtenmacher, C., Kove-
shazi, I., Neumeier, F., Padr
´
on-Laso, V., Jonke,
ˇ
Z.,
Saravi, B., Vollmer, M., Vollmer, A., Hoffmann, J.,
Engel, M., Ristow, O., and Freudlsperger, C. (2021).
Deep learning on oral squamous cell carcinoma ex
vivo fluorescent confocal microscopy data: A feasi-
bility study. Journal of Clinical Medicine, 10(22).
Simonyan, K. and Zisserman, A. (2015). Very deep con-
volutional networks for large-scale image recognition.
In Bengio, Y. and LeCun, Y., editors, 3rd Interna-
tional Conference on Learning Representations, ICLR
2015, San Diego, CA, USA, May 7-9, 2015, Confer-
ence Track Proceedings.
Soni, A., Sethy, P., Dewangan, A., Nanthaamornphong, A.,
Behera, S. K., and Devi, B. (2024). Enhancing oral
squamous cell carcinoma detection: a novel approach
using improved efficientnet architecture. BMC Oral
Health 24, 24:601.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wojna,
Z. (2015). Rethinking the inception architecture for
computer vision. 2016 IEEE Conference on Computer
Vision and Pattern Recognition (CVPR), pages 2818–
2826.
Torres, J., Oliveira, J., and Gomes, E. (2022). The us-
age of data augmentation strategies on the detection
of murmur waves in a pcgsignal. In Proceedings of
the 15th International Joint Conference on Biomed-
ical Engineering Systems and Technologies, volume
4:BIOSIGNALS, pages 128–132.
Histopathological Imaging Dataset for Oral Cancer Analysis: A Study with a Data Leakage Warning
817

Vigneswaran, N. and Williams, M. (2014). Epidemiologic
trends in head and neck cancer and aids in diagno-
sis. Oral and Maxillofacial Surgery Clinics of North
America, 26(2):123–141.
Zeiser, F. A., da Costa, C. A., Zonta, T., Marques, N. M. C.,
Roehe, A. V., Moreno, M., and da Rosa Righi, R.
(2020). Segmentation of masses on mammograms us-
ing data augmentation and deep learning. Journal of
Digital Imaging, 33:858–868.
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
818
