
3D Visualization and Interaction for Studying Respiratory Infections
by Exploiting 2D CNN-Derived Attention Maps and Lung Models
Mohamed El Fateh Hadjarsi, Adnan Mustafic
a
, Mahmoud Melkemi
b
, Iyed Dhahri and
Karim Hammoudi
∗ c
Universit
´
e de Haute-Alsace, IRIMAS, Mulhouse, France
Keywords:
Chest X-Ray Analysis, Biomedical Image Analysis, Biomedical Diagnosis, Radiology, Infection Studies.
Abstract:
Nowadays, research activities in the fields of precision health and biomedical image analysis are developing
rapidly. In this context, research work on the analysis of respiratory infections is still extensively investigated.
Few open source systems with the goal of visualizing and manipulating lungs with infections in 3D space are
currently proposed. Such systems could become an important tool in the training of new radiologists. In the
present work, we propose an approach that allows the user to visualize and interact with respiratory infections
in 3D space by exploiting 2D CNN-derived attention maps. The source code will be made publicly available
at https://github.com/Adn-an/3D-Visualization-and-Interaction-for-Studying-Respiratory-Infections-by-Exp
loiting-2D-Attention-Maps.
1 INTRODUCTION AND
MOTIVATION
Nowadays, research activities in the fields of preci-
sion health and biomedical image analysis are rapidly
evolving. Especially since the COVID-19 pandemic,
many researchers have focused their attention on an-
alyzing chest and respiratory related pathologies. In
particular, chest X-ray images have been extensively
used in this area due to their low intrusivity and high
availability (Hammoudi et al., 2021) (Slika et al.,
2024a).
In the literature, few systems which have the goal
of visualizing and manipulating lungs with infec-
tions in 3D space are currently proposed. Some ap-
proaches for visualizing lung infections in 3D exploit
the use of a fully self-contained holographic computer
(Hololens) which is a specialized Augmented Real-
ity (AR) equipment (e.g. (Liu et al., 2024)) or tra-
ditional infection reconstruction methods with static
views (strategy of marching cubes) which are based
on CT scans (e.g. (Hameed et al., 2024)).
In our case, we presently propose an open-source,
cost-effective (headsetless) and lightweight approach
a
https://orcid.org/0009-0002-2658-7017
b
https://orcid.org/0000-0002-9045-9047
c
https://orcid.org/0000-0002-4804-4796
∗
Contact author.
that allows the user to visualize and dynamically in-
teract with respiratory infections in 3D space from
a conventional laptop having a camera and using an
Augmented Reality marker.
Specifically, our AR-based visualization approach
particularly exploits CNN-based 2D features; namely,
attention maps from chest X-rays (queries) in order to
automatically generate semi-realistic lung representa-
tions with associated infections. Our main contribu-
tion is the proposal of a real-time 3D visualization ap-
proach which facilitates the study of lung characteris-
tics such as anatomical and infected regions through
virtual interactions.
Indeed, medical education faces huge difficul-
ties to form students, practitioners, and future radi-
ologists to the interpretation of chest X-rays (Sait
and Tombs, 2021). Through automated and inter-
active visualization systems of organs using laptops
equipped with cameras and augmented reality tech-
nologies (Abu Halimah et al., 2024; Tene et al., 2024;
Jones et al., 2023; Lastrucci et al., 2024), it is thus
possible to allow a large number of learners to dis-
cover characteristics of the human body (location of
organs, shapes of organs, location of recurring infec-
tions) and to improve the understanding of complex
anatomical concepts.
Additionally, we provide a solution that can inte-
grate various levels of visualization of the loaded lung
models. Furthermore, we propose an approach allow-
Hadjarsi, M. F., Mustafic, A., Melkemi, M., Dhahri, I. and Hammoudi, K.
3D Visualization and Interaction for Studying Respiratory Infections by Exploiting 2D CNN-Derived Attention Maps and Lung Models.
DOI: 10.5220/0013383000003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 413-416
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
413
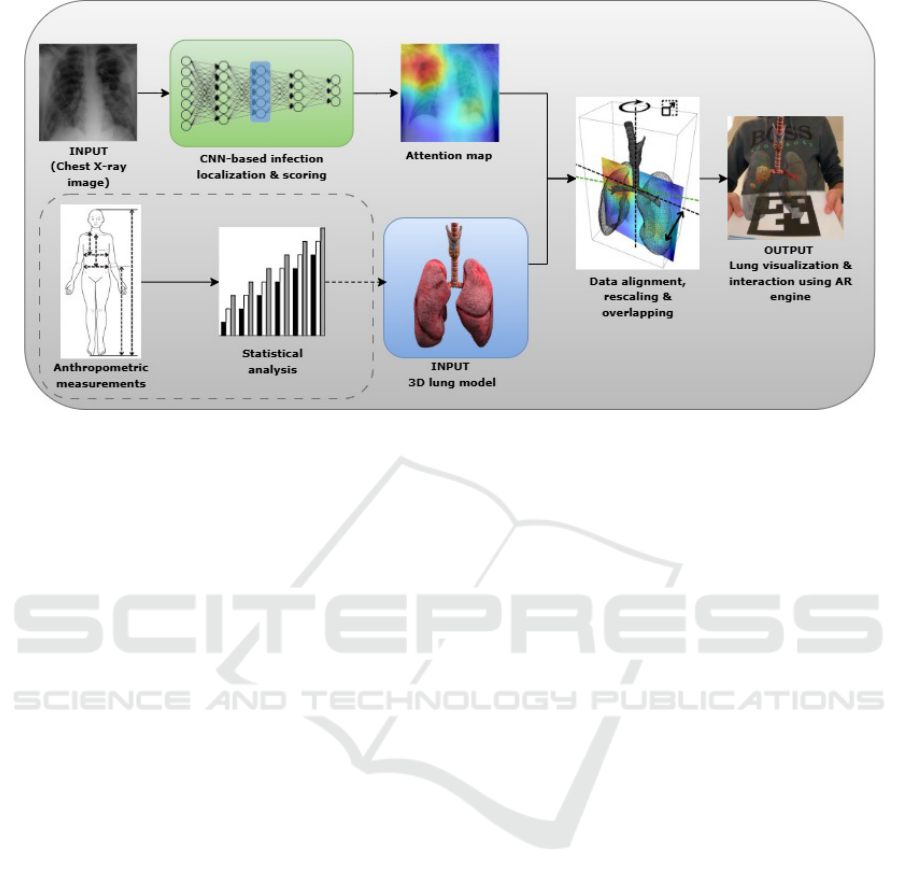
Figure 1: Proposed approach for visualizing and interacting with 3D lung model. The section in dotted lines describes
complementary possibilities to generate a lung model based on anthropometric patient data and population statistics.
ing to load textures with an organic appearance or to
include on the surface of the visualized 3D objects as-
perities including different levels of granularity.
2 PROPOSED APPROACH
2.1 Visualization and Interaction of the
Generated Lung Model
This approach involves exploiting a pre-trained CNN
that aims to localize and quantify the severity of chest
infections from frontal chest X-ray images (Slika
et al., 2024b). Our approach operates in an original
way in the sense that a pre-trained CNN is exploited in
order to extract infection features from a 2D attention
map (saliency map). In this way, the lung infection
areas are potentially represented by automatically an-
alyzing chest X-ray images (initial input). The lungs
are represented by using a realistic 3D lung model
which is the second input.
Then, the 3D lung model is aligned in order to get
the lung frontally oriented. A rescaling step is per-
formed in order to overlap the 2D attention map ac-
cording to the bounding box of the realistic 3D lung
model. The attention map is positioned at the center
of the 3D lung model. We emphasize that the vol-
ume of the lung model can be adjusted according to
the characteristics of people, see the section in dotted
lines in Figure 1, which are provided by the meta-
data associated with the query chest X-ray (statistical
information of the patient e.g., patient gender, size,
height, weight (Sharma and Goodwin, 2006)).
We extract the areas of interest by using estimated
infection severity, represented by high intensities in
the attention map, and their positions. These ex-
tracted infection areas are then represented by spheres
of which dimensions are weighted according to the
severity of the infection, while ensuring that they fit
into the lung model. The resulting 3D lung model
is then loaded into an augmented reality visualizer as
shown in Figure 1. The opacity of the lungs or as-
sociated infections is tuned by exploiting an alpha-
blending approach (e.g. (Friederichs et al., 2021)).
2.2 Enhancement of the Generated
Lung Model
One of the key components of the visualization
method lies in the 3D model itself. The process of
finding, modifying, and enhancing the model has re-
quired a rendering tool and a high-end graphics card.
It also explores the benefits of baking details into
a diffuse map. After finding a 3D lung model on-
line, 3D art techniques are utilized to re-mesh and re-
model such as polygon decimation and shape sculpt-
ing. Then the model is re-textured and re-rendered;
and finally all the textures are correlated to one diffuse
map. That’s mostly aimed at enhancing the realism of
the model, making it faster and easier for graphical
handling.
To bake all these different maps into one, a com-
puter graphics renderer is applied in order to produce
a high-fidelity scene including the subsurface scat-
tering, lighting, bump, displacement, reflections, and
specular maps managed through one single KD map
BIOIMAGING 2025 - 12th International Conference on Bioimaging
414
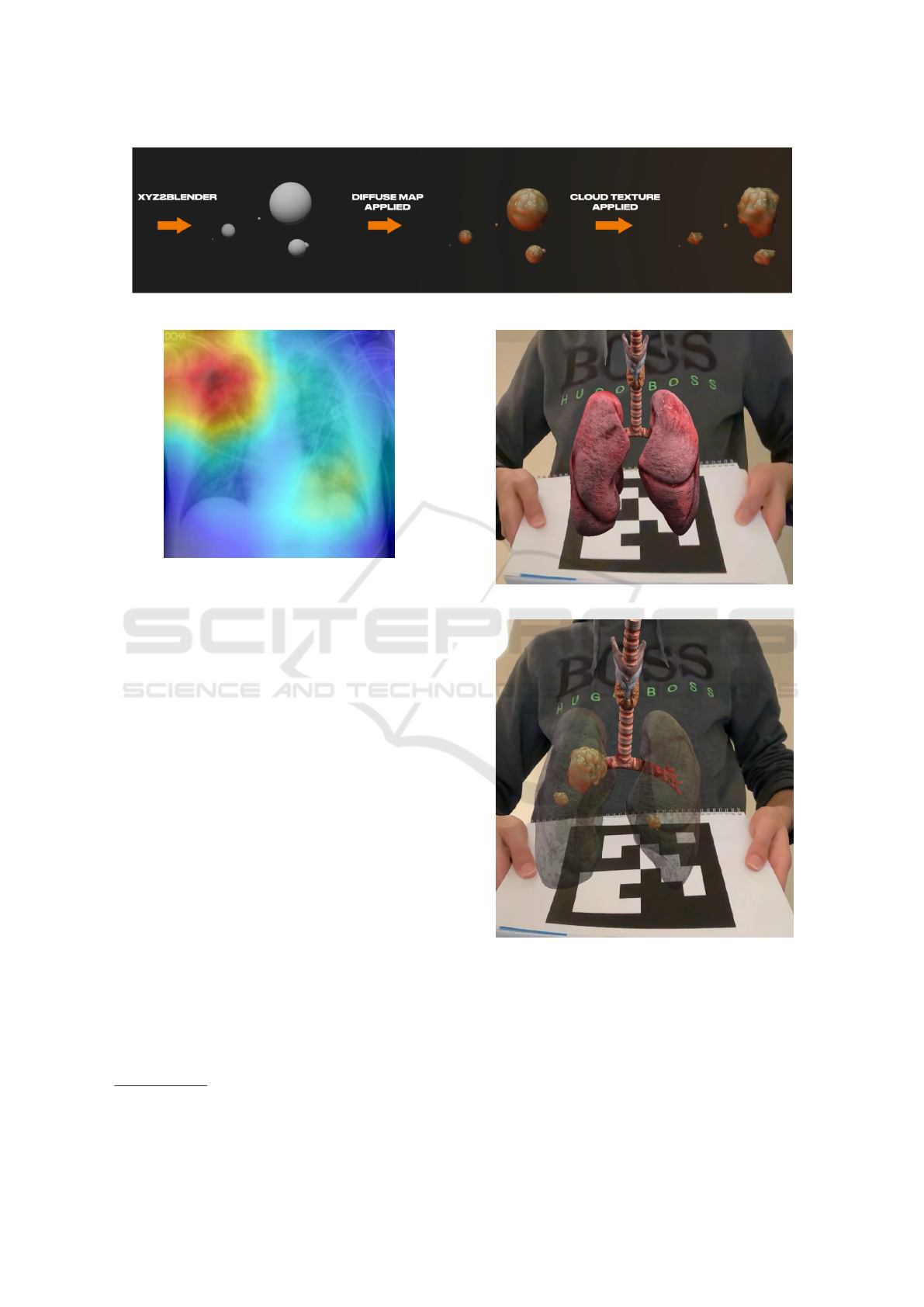
Figure 2: Enhanced infection representations by varying model rendering and surface asperities.
Figure 3: Attention map derived from the CNN architec-
tures which is used for scoring infections (Slika et al.,
2024b); e.g. potential lung infection areas (Misra, 2021).
image, e.g. diffuse map (Yu et al., 2023).
An example of enhancements for our infection
representation is illustrated in Figure 2. As can be
seen, the infected areas can be represented by a sphere
that is scaled according to the localized attention map
intensities. For a realistic visualization, the appear-
ance of the infections can also be enhanced by map-
ping textures over the generated spheres and/or by
modifying the surface of the spheres with asperities.
3 EXPERIMENTAL RESULTS
AND EVALUATION
The approach is based on a visualization model
(Qing, 2022), which is capable of loading a 3D ob-
ject and displaying it on an Augmented Reality en-
gine (ArUco
1
(Garrido-Jurado et al., 2015)(Romero-
Ramirez et al., 2018)) while exploiting computer vi-
sion functionalities (OpenCV
2
). In particular, a cal-
ibration chessboard is used to calibrate the model’s
parameters associated with the camera. Our approach
uses a generic lung model available online (neshal-
lAds, 2022).
1
Aruco: Augmented Reality University Cordoba
2
OpenCV: Open Computer Vision
(a) Opaque lung.
(b) Translucent lung with generated infection areas.
Figure 4: AR results of the lung model with and without
transparency.
Figure 3 displays the input chest X-ray image, ac-
companied by its attention map which highlights re-
gions with potential anomalies (red and yellow re-
gions of the heatmap). In Figure 4a, the initial 3D
lung model is shown on top of the marker, which is
3D Visualization and Interaction for Studying Respiratory Infections by Exploiting 2D CNN-Derived Attention Maps and Lung Models
415

generated from ArUco. Figure 4b shows the views
resulting from the exploitation of the data observed
through Figures 3 and 4a with the lungs shown with a
transparency parameter.
The lung and its features can be visualized with
varied representation levels in the sense that we can
observe the lung without infections, only infections,
or a mix of both the lungs and infections. The ob-
ject rendering has been computed from an RTX 3080
graphics card. The interaction with loaded lung mod-
els operates in real time (approximately 25 frames per
second).
4 CONCLUSIONS AND FUTURE
WORKS
In this paper, we propose a concrete and effective tool
to visualize a 3D lung model and control its opacity
to inspect internal infections in the lung by automat-
ically processing 2D attention maps extracted from a
CNN pre-trained on frontal chest X-ray images. All
features of this analysis tool work in real time, which
shows its usefulness in facilitating chest X-ray studies
and interpretations to physicians, practitioners, and
future radiologists. Moreover, our approach can be
generalized to visualize other organs.
In future work, we aim to better localize the lung
infection in terms of depth by combining lateral with
frontal images of the lung. This pair of images should
give us more information about the shape, depth,
and location of the infection in order to accurately
diagnose the patient and improve patient care.
REFERENCES
Abu Halimah, J., Mojiri, M. E., Ali, A. A., et al. (2024).
Assessing the impact of augmented reality on surgi-
cal skills training for medical students: A systematic
review. Cureus, 16(10):e71221.
Friederichs, F., Eisemann, M., and Eisemann, E. (2021).
Layered weighted blended order-independent trans-
parency. In Proceedings of Graphics Interface 2021,
GI 2021, pages 196 – 202. Canadian Information Pro-
cessing Society.
Garrido-Jurado, S., Mu
˜
noz-Salinas, R., Madrid-Cuevas, F.,
and Medina-Carnicer, R. (2015). Generation of fidu-
cial marker dictionaries using mixed integer linear
programming. Pattern Recognition, 51.
Hameed, H. K., Alazawi, A., Humadi, A. F., and Jameel,
H. F. (2024). Three-dimensional visualization of lung
corona viral infection region-based reconstruction of
computed tomography staked volumetric data using
marching cubes strategy. AIP Conference Proceed-
ings, 3051(1):040019.
Hammoudi, K., Benhabiles, H., Melkemi, M., Dornaika, F.,
Arganda-Carreras, I., Collard, D., and Scherpereel, A.
(2021). Deep learning on chest x-ray images to detect
and evaluate pneumonia cases at the era of COVID-
19. J. Medical Syst., 45(7):75.
Jones, D., Galvez, R., Evans, D., Hazelton, M., Rossiter,
R., Irwin, P., Micalos, P. S., Logan, P., Rose, L., and
Fealy, S. (2023). The integration and application of
extended reality (xr) technologies within the general
practice primary medical care setting: A systematic
review. Virtual Worlds, 2(4):359–373.
Lastrucci, A., Wandael, Y., Barra, A., Ricci, R., Maccioni,
G., Pirrera, A., and Giansanti, D. (2024). Exploring
augmented reality integration in diagnostic imaging:
Myth or reality? Diagnostics, 14(13):1333.
Liu, J., Lyu, L., Chai, S., Huang, H., Wang, F., Tateyama,
T., Lin, L., and Chen, Y. (2024). Augmented reality
visualization and quantification of covid-19 infections
in the lungs. Electronics, 13(6).
Misra, P. (2021). xrays-and-gradcam. https://github.com/p
riyavrat-misra/xrays-and-gradcam. Accessed: 2023-
10-28.
neshallAds (2022). Realistic human lungs. https://sketchfa
b.com/3d-models/realistic-human-lungs-ce09f4099
a68467880f46e61eb9a3531. Accessed: 2024-12-16.
Qing, B. (2022). Opencv ar. https://github.com/BryceQing
/OPENCV AR. Accessed: 2023-10-28.
Romero-Ramirez, F., Mu
˜
noz-Salinas, R., and Medina-
Carnicer, R. (2018). Speeded up detection of squared
fiducial markers. Image and Vision Computing, 76.
Sait, S. and Tombs, M. (2021). Teaching medical students
how to interpret chest x-rays: The design and develop-
ment of an e-learning resource. Advances in Medical
Education and Practice, 12:123–132.
Sharma, G. and Goodwin, J. (2006). Effect of aging on res-
piratory system physiology and immunology. Clin-
ical Interventions in Aging, 1(3):253–260. PMID:
18046878.
Slika, B., Dornaika, F., and Hammoudi, K. (2024a). Multi-
score prediction for lung infection severity in chest x-
ray images. IEEE Transactions on Emerging Topics
in Computational Intelligence, pages 1–7.
Slika, B., Dornaika, F., Merdji, H., and Hammoudi, K.
(2024b). Lung pneumonia severity scoring in chest
x-ray images using transformers. Medical & Biologi-
cal Engineering & Computing, 62(8):2389–2407.
Tene, T., Marcatoma Tixi, J. A., Palacios Robalino, M. d. L.,
Mendoza Salazar, M. J., Vacacela Gomez, C., and
Bellucci, S. (2024). Integrating immersive technolo-
gies with stem education: a systematic review. Fron-
tiers in Education, 9.
Yu, X., Dai, P., Li, W., Ma, L., Liu, Z., and Qi, X. (2023).
Texture generation on 3d meshes with point-uv diffu-
sion. In Proceedings of the IEEE/CVF International
Conference on Computer Vision, pages 4206–4216.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
416
