
Leveraging Attention Mechanisms for Interpretable Human
Embryo Image Segmentation
Wided Souid Miled
1,2
and Nozha Chakroun
3
1
LIMTIC Laboratory, Higher Institute of Computer Science, University of Tunis El-Manar, Ariana, Tunisia
2
National Institute of Applied Science and Technology, University of Carthage, Centre Urbain Nord, Tunisia
3
University of Medicine of Tunis, Lab. of Reproductive Biology and Cytogenetic, Aziza Othmana Hospital, Tunisia
Keywords:
In Vitro Fertilization, Embryo Selection, Blastocyst Segmentation, Attention Mechanisms, Deep Learning,
Explainability.
Abstract:
In-vitro Fertilization (IVF) is a widely used assisted reproductive technology where embryos are cultured un-
der controlled laboratory conditions. The selection of a high-quality blastocyst, typically reached five days
after fertilization, is crucial to the success of the IVF procedure. Therefore, evaluating embryo quality at this
stage is essential to optimize IVF outcomes. Advances in neural network architectures, particularly Convo-
lutional Neural Networks (CNNs), have enhanced decision-making in IVF. However, ensuring both accuracy
and interpretability in these models remains a challenge. This paper focuses on improving human blastocyst
segmentation by combining channel attention mechanisms with a ResNet50 model within an encoder-decoder
architecture. The method accurately identifies key blastocyst components such as inner cell mass (ICM), tro-
phectoderm (TE), and zona pellucida (ZP). Our approach was validated on a publicly available human embryo
dataset, achieving Intersection over Union (IoU) scores of 83.09% for ICM, 86.87% for ZP, and 81.1% for
TE, outperforming current state-of-the-art methods. These results demonstrate the potential of deep learning
to improve both accuracy and interpretability in embryo quality assessment.
1 INTRODUCTION
In vitro fertilization (IVF) is one of the most widely
used and effective forms of assisted reproductive tech-
nology (ART) that helps couples facing fertility chal-
lenges conceive a child. Since 1978, more than 9 mil-
lion babies have been born through IVF, and approx-
imately 6% of couples experiencing infertility turn to
this procedure (Kuhnt and Passet-Wittig, 2022). The
IVF process involves several critical stages. First, ma-
ture eggs are retrieved from the ovaries and manually
combined with sperm in a controlled environment for
fertilization. The resulting fertilized egg, now called
embryo, undergoes a series of developmental phases.
Initially, the male and female pronuclei appear and
then disappear, followed by the cleavage stage, where
the single cell divides into multiple cells. Four days
after fertilization, the embryo compacts, reaching the
morula stage, and by the fifth day, the embryo devel-
ops into a blastocyst.
One of the most critical steps in this complex pro-
cess is embryo selection, which aims to identify the
healthiest embryo with the highest likelihood of re-
sulting in the birth of a healthy baby. Embryo quality
is considered a key predictor of success in IVF cy-
cles. Numerous studies have demonstrated a strong
correlation between embryo morphology, implanta-
tion rates, and clinical pregnancy outcomes (Shulman
et al., 1993; Dennis et al., 2006). Consequently, se-
lecting a high-quality embryo significantly increases
the potential for a successful pregnancy. A notewor-
thy advancement in the field of IVF is the introduction
of time-lapse imaging incubators (TLI). This innova-
tive technology has transformed the embryo selection
process by providing a dynamic, real-time view of
embryonic development. TLI systems capture images
of each embryo at regular intervals and compile them
into a time-lapse video, offering dynamic insight into
embryonic development in vitro without disturbing
the stable culture conditions (Kovacs, 2016; Good-
man et al., 2016). Using these time-lapse videos,
embryologists grade blastocysts and identify the em-
bryos with the greatest pregnancy potential for trans-
fer to the woman’s uterus.
872
Miled, W. S. and Chakroun, N.
Leveraging Attention Mechanisms for Interpretable Human Embryo Image Segmentation.
DOI: 10.5220/0013383200003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 2, pages 872-879
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

At the blastocyst stage, the embryo consists of two
main inner regions, the inner cell mass (ICM), and
the trophectoderm epithelium (TE). These regions are
surrounded by an outer membrane, the zona pellu-
cida (ZP). Both ICM and TE are considered key mor-
phological parameters for assessing embryo viability.
Therefore, evaluating the quality of these regions is
essential to determine embryonic potential. The most
commonly employed grading system for blastocyst
morphology is that of Gardner et al. (Gardner and
Schoolcraft, 1999). According to this system, three
crucial parameters are used to predict successful preg-
nancy outcomes: the degree of blastocoel cavity ex-
pansion relative to the zona pellucida, the compact-
ness of the inner cell mass, and the density of the
trophectoderm. Accurate measurement of these pa-
rameters requires segmenting human blastocyst im-
ages to delineate the three regions. Manual identifica-
tion of these regions is challenging, time-consuming,
and subjective, often leading to variability between
experts. Recently, many efforts have been made to
automate embryo segmentation, utilizing both classi-
cal and deep learning-based approaches (Filho et al.,
2012; Saeedi et al., 2017; Rad et al., 2019; Muham-
mad et al., 2022). Despite its importance, embryo
segmentation still presents several challenges due to
significant variability in the scale, shape, position, and
orientation of blastocyst components. Convolutional
Neural Networks (CNNs) have emerged as state-of-
the-art tools in medical image segmentation, offer-
ing remarkable performance in various applications.
However, conventional CNN architectures face cer-
tain limitations when applied to this task. One key is-
sue is their limited spatial awareness, particularly for
flexible structures with varying shapes and positions,
as CNNs rely on shared weights in convolutional lay-
ers. This limitation can affect accurate segmenta-
tion, especially when dealing with complex, non-rigid
structures such as the inner cell mass, trophectoderm,
and zona pellucida. Moreover, the high dimensional-
ity of feature maps in CNNs can lead to redundancy,
reducing computational efficiency and increasing pro-
cessing time. Another significant challenge is the lack
of interpretability in CNN decision-making. While
CNNs excel at extracting features, their black-box na-
ture makes it difficult to understand why certain deci-
sions are made. This lack of explainability is a ma-
jor drawback in clinical settings, where transparent
decision-making is crucial for medical professionals
to trust and act on the results.
To address these challenges, this work focuses on
improving human embryo segmentation. As a critical
step in the IVF process, embryo segmentation enables
clinicians to assess embryo viability by delineating
key regions, including Inner Cell Mass, Zona Pellu-
cida, and Trophectoderm. Accurate segmentation of
these regions is essential for identifying embryos with
the highest implantation potential, thereby improving
IVF success rates. Building on recent advancements
in deep learning, this study proposes a novel approach
to tackle the dual challenges of segmentation accu-
racy and model interpretability.
The main contributions of this work are as follows:
• We integrate channel attention mechanisms with
a ResNet50 backbone in an encoder-decoder con-
volutional neural network. This architecture en-
hances the model’s ability to detect key hu-
man blastocyst components while providing in-
terpretable insights into the decision-making pro-
cess.
• The proposed approach is evaluated using a pub-
licly available blastocyst segmentation dataset. Its
performance is compared to state-of-the-art meth-
ods and a baseline U-Net model augmented with
the post-hoc explainability technique Grad-CAM.
• This work addresses the dual challenges of accu-
racy and interpretability in deep learning models
for embryo segmentation. By providing a frame-
work that ensures reliable segmentation while en-
hancing model transparency, it contributes to bet-
ter healthcare outcomes and fosters greater trust
in AI-driven medical solutions.
2 EXPLAINABLE AI FOR IMAGE
SEGMENTATION
Segmentation involves the automatic identification
and delineation of specific objects or regions within
an image. The integration of explainable artificial in-
telligence (XAI) into image segmentation is gaining
attention for its potential to enhance the fairness and
reliability of AI models, particularly in sensitive do-
mains like healthcare (Saranya and Subhashini, 2023;
Gunashekar et al., 2022). In clinical applications, in-
terpretable segmentation models are essential to build
trust, improve communication with patients, and sup-
port iterative refinements to achieve more precise and
accurate results. This section explores XAI methods
designed to clarify the decision-making processes of
segmentation models, a critical step toward enhancing
their applicability in clinical contexts and advancing
research efforts.
Leveraging Attention Mechanisms for Interpretable Human Embryo Image Segmentation
873

2.1 Ad-Hoc Explainability Methods
Ad-hoc explainability methods are external tech-
niques applied to pre-existing AI models to provide
insight into their decision-making processes. These
methods are particularly valuable for complex mod-
els, such as deep learning architectures, which in-
herently lack transparency. Commonly used ad hoc
techniques for image segmentation include SHapley
Additive exPlanations (SHAP), Local Interpretable
Model-agnostic Explanations (LIME), and Gradient-
weighted Class Activation Mapping (Grad-CAM).
SHAP assigns contribution scores to input features,
quantifying their influence on the model’s predictions.
By leveraging game theory, SHAP ensures fair attri-
bution of prediction credit, providing a detailed un-
derstanding of the importance of the feature. LIME
generates interpretable surrogate models to approxi-
mate the behavior of a complex model around a spe-
cific prediction. This helps explain individual predic-
tions, especially in ambiguous or error-prone cases.
The Grad-CAM approach (Selvaraju et al., 2017;
Vinogradova et al., 2020) utilizes gradients from the
final convolutional layer to highlight image regions
that most influence the model’s predictions, provid-
ing intuitive visual explanations. Its enhanced vari-
ant, Grad-CAM++, incorporates information from all
convolutional layers, refining the localization of dis-
criminative regions and delivering more precise ex-
planations.
2.2 Self-Explainable Methods
Unlike ad-hoc methods, self-explainable models are
designed with interpretability as an integral part of
their architecture. These models inherently inte-
grate explainability into their structure, making their
decision-making processes clear. One prominent ex-
ample is the ProtoSeg method, proposed by Sacha
et al. (Sacha et al., 2023), which uses prototype-
based learning to assign pixels to classes by compar-
ing image regions to prototypes, which are character-
istic examples of each class. This approach naturally
enhances interpretability, as segmentation decisions
are explicitly linked to these prototypes. Attention-
based models leverage attention mechanisms to focus
on relevant regions within an image during segmen-
tation (Gu et al., 2020; Zhao et al., 2021). By as-
signing importance scores to features, they generate
interpretable attention maps, simultaneously improv-
ing segmentation accuracy and transparency.
The choice between ad hoc and self-explainable
methods depends on the specific requirements of the
applications. Ad-hoc methods are suitable for provid-
ing detailed and flexible analysis, making them ideal
for in-depth evaluations of existing models. How-
ever, they often require technical expertise and com-
putational resources. In contrast, self-explainable
methods prioritize intuitive interpretation and com-
putational efficiency, making them well-suited for
resource-constrained environments or users with lim-
ited technical expertise.
3 METHODOLOGY
3.1 Model Architecture
3.1.1 Overview
The proposed Residual Attention Network (ResA-
Net), inspired by the method introduced in (Gu et al.,
2020), integrates comprehensive attention mecha-
nisms for efficient and interpretable image segmen-
tation. While retaining the general structure of the
original model, we enhanced its backbone by replac-
ing it with ResNet50 to leverage its superior feature
extraction capabilities. Specialized attention modules
are incorporated to guide segmentation across spatial,
channel, and scale dimensions simultaneously. The
architecture comprises four spatial attention modules
(SA1-SA4), four channel attention modules (CA1-
CA4), and a scale attention module (LA), each con-
tributing to precise and context-aware segmentation.
ResA-Net adopts an encoder-decoder architec-
ture. The encoder processes input images through
convolutional layers and max-pooling operations, re-
ducing spatial dimensions while capturing high-level
and multi-scale features. These features are passed
to a central layer, bridging the encoder and decoder,
before being fed into the decoder to reconstruct the
segmentation output. The decoder employs bilinear
interpolation and concatenation to upsample feature
maps to the original resolution while effectively com-
bining information from multiple scales. Finally, a
1×1 convolutional layer reduces feature map dimen-
sions, and a class-specific convolutional layer gener-
ates probability maps for each segmentation class. A
softmax layer converts these probabilities into the fi-
nal segmentation output by assigning each pixel to the
class with the highest likelihood. An overview of the
proposed method is presented in Figure 1.
While achieving accurate segmentation is essen-
tial, our focus extends beyond the segmentation task
to understanding the model’s decision-making pro-
cess. To this end, we emphasize the visualization of
attention maps generated by ResA-Net, which high-
light influential regions in the input image. These
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
874
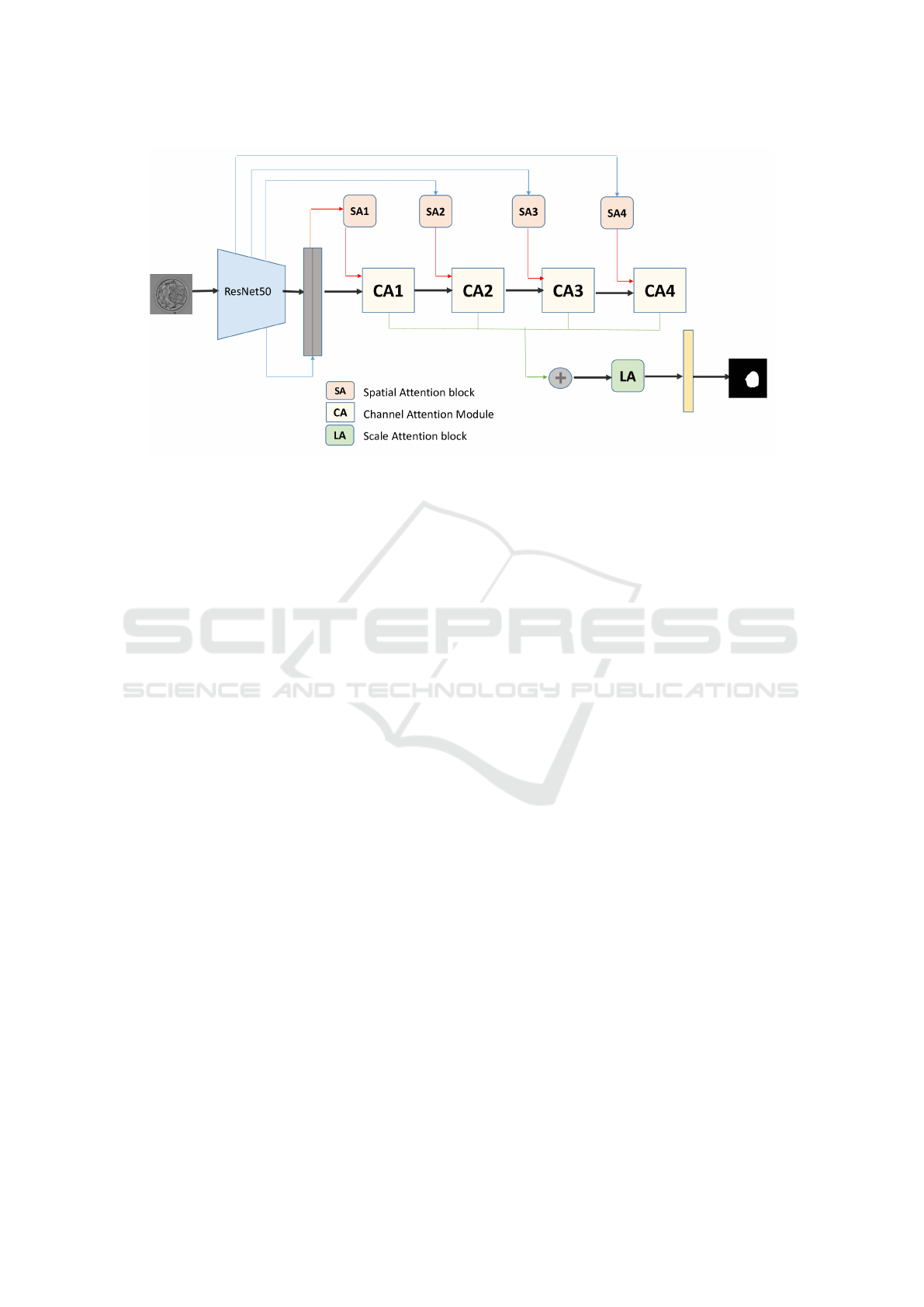
Figure 1: Overview of the proposed ResA-Net model.
visualizations offer valuable insights into how the
model makes its decisions, enhancing transparency
and interpretability by clearly showcasing the regions
and features that guide its segmentation outcomes.
3.1.2 Attention Mechanisms in ResA-Net
ResA-Net employs three attention mechanisms Spa-
tial Attention Modules (SA), Channel Attention Mod-
ules (CA), and a Scale Attention Module (LA).
Spatial Attention: Inspired by the non-local net-
work and Attention Gates (AG), ResA-Net uses four
spatial attention blocks (SA1-SA4) to learn atten-
tion maps across different resolution levels. These
maps emphasize relevant regions of the input image
while suppressing noise, enhancing segmentation ac-
curacy. At the lowest resolution level (SA1), a non-
local block captures global context, while at higher
levels (SA2-SA4), attention gates focus on key areas
within the image. Low-level spatial attention features
from the encoder are concatenated with high-level de-
coder features to enhance segmentation through com-
plementary information.
Channel Attention: Channel attention modules au-
tomatically identify and amplify relevant feature
channels while suppressing irrelevant ones. This
mechanism ensures that the model focuses on seman-
tically meaningful features crucial for accurate seg-
mentation.
Scale Attention: To handle variations in object
scale, the scale attention module assigns adaptive
weights to features at different scales. This allows
the network to prioritize features most relevant to the
specific image, improving segmentation performance.
By combining these mechanisms, ResA-Net not only
achieves high segmentation accuracy but also pro-
vides interpretable outputs, offering insights into the
decision-making process and enabling its application
in sensitive fields such as healthcare.
3.2 Dataset
The dataset employed in this paper for segmenting
human embryo images is the unique publicly avail-
able dataset introduced by Saaedi et al. (Saeedi et al.,
2017). It comprises 235 blastocyst images from pa-
tients treated at the Pacific Center of Reproductive
Medicine in Canada between 2012 and 2016. Each
image was manually annotated by expert embryolo-
gists and provided with ground truth binary masks
relating to ICM, TE, and ZP regions. Using this
dataset, we employed the proposed model architec-
ture to train three distinct configurations, each fo-
cused on segmenting a specific region of interest. This
work adopts a single-class segmentation approach to
individually identify the ZP, ICM, and TE regions
within blastocyst images. By generating explicit seg-
mentation labels, the model facilitates both accurate
segmentation and enhanced interpretability analysis.
4 EXPERIMENTAL RESULTS
4.1 Implementation Details
A consistent preprocessing pipeline was applied to all
three one-class segmentation configurations to ensure
uniformity and efficiency. All images and their corre-
Leveraging Attention Mechanisms for Interpretable Human Embryo Image Segmentation
875

sponding ground truth masks were resized to a fixed
resolution of 224× 300, to ensure consistent input di-
mensions for the model. Both the images and masks
were then converted to .npy format to facilitate effi-
cient storage and handling during training.
To manage the dataset, we implemented a custom
dataset class using PyTorch’s Dataset module. This
class was specifically designed to pair image sam-
ples with their corresponding labels, with each dataset
entry represented as tensors loaded from the prepro-
cessed .npy files. The dataset was divided into train-
ing (85%), and testing (15%) subsets, ensuring a bal-
anced evaluation of the model’s performance. Data
loading was performed using PyTorch’s DataLoader,
enabling efficient batching and streamlined access to
the dataset. A batch size of 16 images was selected to
balance memory constraints and training efficiency.
The Adam optimizer was employed with a learning
rate of 10
−4
and a weight decay of 10
−8
, to opti-
mize the model parameters. The Dice loss function
was utilized to evaluate segmentation performance,
as it effectively handles the class imbalance inherent
in medical imaging datasets. Each configuration was
trained for 100 epochs to ensure convergence and op-
timal performance. Training was conducted locally
on a desktop PC equipped with an NVIDIA GeForce
GTX 1650 graphics card, providing sufficient compu-
tational power for the segmentation task.
4.2 Evaluation
For the performance evaluation of the proposed
model, we used the Intersection over Union (IoU),
also known as the Jaccard Index, and the F1 score.
The Jaccard Index is the ratio of the intersection to
the union of the segmentation result and the ground
truth. The F1 score is defined as twice the area
of overlap between the segmentation result and the
ground truth, divided by the sum of the areas of
both. Table 1 presents a quantitative comparison
of the segmentation performance between the pro-
posed method, the baseline U-Net model and recent
state-of-the art methods. U-Net (Ronneberger et al.,
2015) has demonstrated its effectivenes in segmenting
anatomical structures across various imaging modali-
ties, including MRI, CT scans, and microscopy. Blas-
Net (Rad et al., 2019) enhances segmentation by in-
corporating multi-scale global contextual information
through a cascaded atrous pyramid pooling module
and reproducing feature resolution using dense pro-
gressive sub-pixel upsampling. MASS-Net (Muham-
mad et al., 2022) leverages depth-wise concatenation
to combine spatial information at multiple scales, en-
abling the simultaneous detection of blastocyst com-
ponents. ECS-Net (Mushtaq et al., 2022) employs
dual streams: base convolutional and depth-wise sep-
arable convolutional blocks, densely concatenated to
enrich features for improved segmentation. FSBS-
Net (Ishaq et al., 2023) introduces feature supple-
mentation across scales and employs ascending chan-
nel convolutional blocks (ACCB) to refine blastocyst
segmentation with minimal computational overhead,
leveraging skip connections to integrate features from
shallow and deep layers.
Based on the Intersection over Union (IoU) met-
ric, the ResA-Net model consistently outperforms
state-of-the-art models in segmenting the ZP and TE
regions. While its performance on the ICM region
is slightly below that of some state-of-the-art mod-
els, ResA-Net achieves outstanding results in terms of
the mean IoU across all three regions. This demon-
strates the model’s overall effectiveness and robust-
ness in segmentation tasks.
4.3 Models Interpretability
4.3.1 Visualization with Grad-CAM for U-Net
To better understand the areas of interest that guide
the U-Net model’s segmentation decisions, we used
Grad-CAM to generate heatmaps for each segmen-
tation class. These heatmaps were then combined
into a single heatmap that highlights the regions the
model focuses on during the segmentation process.
Grad-CAM was applied to the last convolutional layer
of the U-Net model, providing insight into how the
model attends to different regions of the image. For
the ZP region, the heatmap shows clear focus on spe-
cific areas within the embryo’s perimeter, which is
the most straightforward to segment. In contrast, for
the ICM region, the model appears to focus on a sin-
gle, central entity within the embryo. This suggests
that the model identifies a distinct, central structure to
guide segmentation. The Trophectoderm (TE) region,
which has the lowest segmentation metric scores, is
the most challenging to identify. The Grad-CAM
heatmap reveals that the model attempts to identify
multiple entities within the inner perimeter of the em-
bryo, which later converge to form the TE region. Fig-
ure 2 presents examples of the combined Grad-CAM
outputs applied to the final layers of the three U-Net
configurations, illustrating the model’s focus for each
segmentation class.
4.3.2 Visualization of ResA-Net Attention Maps
Figure 3 presents the attention maps generated by the
ResA-Net model for each segmentation configuration
(ICM, ZP, TE), derived from the last image in the fi-
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
876
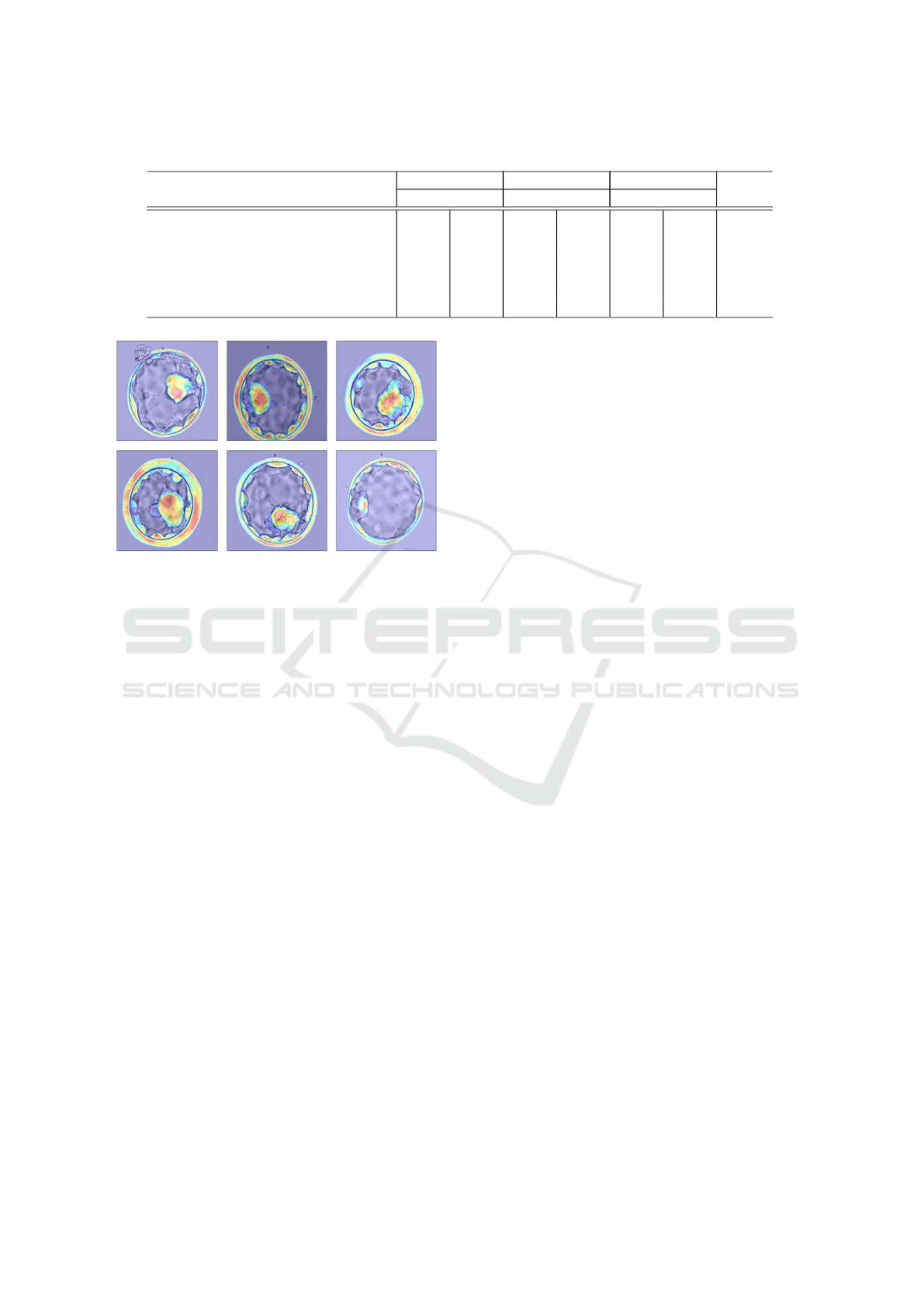
Table 1: Comparative analysis of proposed method with state-of-the-art models in terms of IoU and F1 score.
ZP TE ICM Mean
Model IoU F1 IoU F1 IoU F1 IoU
U-Net Baseline 79.10 88.33 71.66 83.49 76.33 86.57 75.69
BlastNet (Rad et al., 2019) 81.15 - 76.52 - 81.07 - 79.58
MASS-Net (Muhammad et al., 2022) 84.69 - 79.08 - 85.88 - 83.22
ECS-Net (Mushtaq et al., 2022) 85.34 - 78.43 - 85.26 - 83.01
FSBS-Net (Ishaq et al., 2023) 85.80 92.29 80.17 88.90 85.55 92.0 83.84
ResA-Net 86.87 92.72 81.10 88.88 83.09 90.09 83.68
Figure 2: GradCAM heatmaps for U-Net model.
nal batch of the training dataset. These maps offer
valuable insights into the model’s segmentation pro-
cess for each class. In the case of the Zona Pellucida
one-class segmentation configuration, the attention
maps reveal a precise and systematic segmentation
process. The first map clearly delineates the ZP from
its surrounding background, while subsequent maps
progressively refine the segmentation by emphasiz-
ing key feature channels, particularly highlighting the
boundary between the ZP and TE regions. This pro-
gression helps accurately define the contours separat-
ing these regions, showcasing the model’s ability to
focus on the critical features needed for precise seg-
mentation.
For the Inner Cell Mass one-class configuration,
the attention maps demonstrate how the model iden-
tifies the relevant regions within the blastocyst. The
initial map shows the model’s broad understanding of
the entire blastocyst, detecting it as a whole. The sec-
ond map narrows in on the ICM, accurately localizing
this important region. This sequential refinement in-
dicates the model’s capacity to differentiate between
the various components of the blastocyst, enhancing
segmentation precision. However, a slight overlap is
observed between the ICM and TE regions, where
some TE pixels are also highlighted. While this mi-
nor ambiguity does not significantly affect the overall
performance, it underscores the challenges of distin-
guishing adjacent regions.
In contrast, the attention maps for the Trophec-
toderm one-class configuration reveal challenges in
segmentation. Unlike the ZP and ICM models, the
TE model struggles to isolate the TE region. The ini-
tial attention maps fail to clearly differentiate the TE
from the background or the blastocyst structure. De-
spite relatively high segmentation metrics for the TE
region, these scores are lower compared to those for
ZP and ICM, suggesting that this particular input im-
age may not fully represent the model’s performance
across the entire dataset.
5 CONCLUSION
In IVF procedures, accurate segmentation is essen-
tial for assessing embryo viability. By dividing the
embryo into distinct regions, such as the Inner Cell
Mass, Zona Pellucida, and Trophectoderm, clinicians
can better identify embryos suitable for implanta-
tion, ultimately improving the chances of a success-
ful pregnancy. This paper introduces the development
of three one-class segmentation models for embryo
image analysis, incorporating attention mechanisms
with a ResNet50 backbone in an encoder-decoder
architecture. Experimental results demonstrate that
the integration of attention mechanisms enhances the
model’s ability to focus on relevant regions, leading
to improved segmentation performance.
To further enhance segmentation results, future
work could focus on refining the current models by in-
corporating additional contextual information, which
would improve the differentiation of closely related
regions. Additionally, exploring more advanced at-
tention mechanisms and hybrid architectures, com-
bining CNNs with transformer models, could cap-
ture fine-grained details more effectively, ultimately
boosting performance.
Leveraging Attention Mechanisms for Interpretable Human Embryo Image Segmentation
877

Figure 3: Attention Heatmaps Generated by the ResA-Net Model.
REFERENCES
Dennis, S. J., Thomas, M. A., Williams, D. B., and Robins,
J. C. (2006). Embryo morphology score on day 3 is
predictive of implantation and live birth rates. Journal
of assisted reproduction and genetics, 23(4).
Filho, E., Noble, J., Poli, M., Griffiths, T., Emerson, G., and
Wells, D. (2012). A method for semi-automatic grad-
ing of human blastocyst microscope images. Human
reproduction (Oxford, England), 27:2641–2648.
Gardner, D. K. and Schoolcraft, W. B. (1999). Culture and
transfer of human blastocysts. Current Opinion in Ob-
stetrics and Gynaecology, 11(3):307–311.
Goodman, L. R., Goldberg, J., Falcone, T., Austin, C., and
Desai, N. (2016). Does the addition of time-lapse
morphokinetics in the selection of embryos for trans-
fer improve pregnancy rates? a randomized controlled
trial. Journal of Fertility and Sterility, 105(2).
Gu, R., Wang, G., Song, T., Huang, R., Aertsen, M., De-
prest, J., Ourselin, S., Vercauteren, T., and Zhang, S.
(2020). Ca-net: Comprehensive attention convolu-
tional neural networks for explainable medical image
segmentation. IEEE transactions on medical imaging,
40(2):699–711.
Gunashekar, D. D., Bielak, L., H
¨
agele, L., Oerther, B.,
Benndorf, M., Grosu, A., Brox, T., Zamboglou, C.,
and Bock, M. (2022). Explainable ai for cnn-based
prostate tumor segmentation in multi parametric mri
correlated to whole mount histopathology. Radiation
Oncology, 17(1):65.
Ishaq, M., Raza, S., Rehar, H., Abadeen, S., Hussain, D.,
Naqvi, R., and Lee, S. W. (2023). Assisting the human
embryo viability assessment by deep learning for in
vitro fertilization. Mathematics, 11(9):2023.
Kovacs, P. (2016). Time-lapse embryoscopy: Do we have
an efficacious algorithm for embryo selection? Jour-
nal of Reproductive Biotechnology and Fertility, 5.
Kuhnt, A. K. and Passet-Wittig, J. (2022). Families formed
through assisted reproductive technology: Causes, ex-
periences, and consequences in an international con-
text. In Reprod Biomed Soc Online, number 14, pages
289–296.
Muhammad, A., Adnan, H., Woon, C. S., Hwan, K. Y.,
and Ryoung, P. K. (2022). Human blastocyst com-
ponents detection using multiscale aggregation se-
mantic segmentation network for embryonic analysis.
Biomedicines, 10(7):1717.
Mushtaq, A., Mumtaz, M., Raza, A., Salem, N., and Yasir,
M. N. (2022). Artificial intelligence-based detection
of human embryo components for assisted reproduc-
tion by in vitro fertilization. Sensors, 22(19):7418.
Rad, R. M., Saeedi, P., Au, J., and Havelock, J. (2019).
Blast-net: Semantic segmentation of human blasto-
cyst components via cascaded atrous pyramid and
dense progressive upsampling. In IEEE International
Conference on Image Processing (ICIP), pages 1865–
1869.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical image computing and
computer-assisted intervention–MICCAI 2015: 18th
international conference, Munich, Germany, October
5-9, 2015, proceedings, part III 18, pages 234–241.
Springer.
Sacha, M., Rymarczyk, D., Struski, L., Tabor, J., and
Zieli
´
nski, B. (2023). Protoseg: Interpretable semantic
segmentation with prototypical parts. In Proceedings
of the IEEE/CVF Winter Conference on Applications
of Computer Vision, pages 1481–1492.
Saeedi, P., Yee, D., Au, J., and Havelock, J. (2017). Auto-
matic identification of human blastocyst components
via texture. IEEE Transactions on Biomedical Engi-
neering, 64(12):2968–2978.
Saranya, A. and Subhashini, R. (2023). A systematic re-
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
878

view of explainable artificial intelligence models and
applications: Recent developments and future trends.
Decision analytics journal, page 100230.
Selvaraju, R., Cogswell, M., Das, A., Vedantam, R., Parikh,
D., and Batra, D. (2017). Grad-cam: Visual explana-
tions from deep networks via gradient-based localiza-
tion. In Proceedings of the IEEE international confer-
ence on computer vision, pages 618–626.
Shulman, A., Ben-Nun, I., Y. Ghetler, H. K., Shilon, M.,
and Beyth, Y. (1993). Relationship between embryo
morphology and implantation rate after in vitro fertil-
ization treatment in conception cycles. Fertility and
sterility, 60(1).
Vinogradova, K., Dibrov, A., and Myers, G. (2020).
Towards interpretable semantic segmentation via
gradient-weighted class activation mapping (student
abstract). Proceedings of the AAAI Conference on Ar-
tificial Intelligence, 34(10):13943–13944.
Zhao, Q., Liu, J., Li, Y., and Zhang, H. (2021). Seman-
tic segmentation with attention mechanism for remote
sensing images. IEEE Transactions on Geoscience
and Remote Sensing, PP:1–13.
Leveraging Attention Mechanisms for Interpretable Human Embryo Image Segmentation
879
