
Quantum-Efficient Kernel Target Alignment
Rodrigo Coelho
1
, Georg Kruse
1,2
and Andreas Rosskopf
1
1
Fraunhofer IISB, Erlangen, Germany
2
Technical University Munich, Germany
{rodrigo.coelho, georg.kruse, andreas.rosskopf}@iisb.fraunhofer.de
Keywords:
Quantum Machine Learning, Quantum Kernels, Kernel Target Alignment.
Abstract:
In recent years, quantum computers have emerged as promising candidates for implementing kernels. Quan-
tum Embedding Kernels embed data points into quantum states and calculate their inner product in a high-
dimensional Hilbert Space by computing the overlap between the resulting quantum states. Variational Quan-
tum Circuits (VQCs) are typically used for this end, with Kernel Target Alignment (KTA) as cost function. The
optimized kernels can then be deployed in Support Vector Machines (SVMs) for classification tasks. However,
both classical and quantum SVMs scale poorly with increasing dataset sizes. This issue is exacerbated in quan-
tum kernel methods, as each inner product requires a quantum circuit execution. In this paper, we investigate
KTA-trained quantum embedding kernels and employ a low-rank matrix approximation, the Nystr
¨
om method,
to reduce the quantum circuit executions needed to construct the Kernel Matrix. We empirically evaluate the
performance of our approach across various datasets, focusing on the accuracy of the resulting SVM and the
reduction in quantum circuit executions. Additionally, we examine and compare the robustness of our model
under different noise types, particularly coherent and depolarizing noise.
1 INTRODUCTION
Quantum computers may potentially solve certain
problems faster than classical computers. However,
in the Noisy Intermediate Scale Quantum (NISQ) era,
we are limited in the algorithms that quantum com-
puters can implement (Preskill, 2018). Consequently,
algorithms that theoretically offer advantages over the
best-known classical methods, such as Shor’s (Shor,
1999) and Grover’s (Grover, 1996) algorithms, cannot
yet be implemented to tackle problems of industrial
significance. In the NISQ era, quantum computing
has focused on Variational Quantum Circuits (VQCs).
These circuits rely on free parameters that are iter-
atively updated by a classical optimizer. VQCs are
suitable for NISQ devices due to their low require-
ments in both number of qubits and circuit depth,
which mitigates noise effects (Cerezo et al., 2021).
Typically used as function approximators, VQCs are
the quantum analog of Neural Networks (NNs), as
both are black-box models that depend on parame-
ters iteratively adjusted to minimize a cost function
(Abbas et al., 2021). Given their potential, VQCs
have been extensively applied in machine learning
and form a significant component of Quantum Ma-
chine Learning (QML). Notable examples include
the Quantum Approximate Optimization Algorithm
(QAOA) (Farhi et al., 2014) for solving combinato-
rial optimization problems, the Variational Quantum
Eigensolver (VQE) (Kandala et al., 2017) for finding
ground states of Hamiltonians, and their application
in both supervised (Schuld, 2018) and reinforcement
(Skolik et al., 2022) learning.
On the classical side, kernel methods are one of
the cornerstones of machine learning, known for their
effectiveness in handling non-linear data by using im-
plicit feature spaces. These methods map input data
into a higher-dimensional space where linear separa-
tion is possible, facilitated by the kernel trick, which
computes inner products in this space without explic-
itly performing the transformation. Support Vector
Machines (SVMs) are a prime example, leveraging
kernel methods to find optimal decision boundaries
for classification tasks (Hearst et al., 1998). Even
though we’ll focus on SVMs throughout this work,
kernel methods can be applied to many tasks beyond
classification, ranging from regression (Drucker et al.,
1996) to clustering (Dhillon et al., 2004).
In recent years, interest in exploring how quan-
tum computing can enhance kernel methods has in-
creased. Quantum computers, can naturally pro-
cess high-dimensional spaces, providing a promising
Coelho, R., Kruse, G. and Rosskopf, A.
Quantum-Efficient Kernel Target Alignment.
DOI: 10.5220/0013391500003890
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 1, pages 763-772
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
763

platform for implementing kernels. For this reason,
they have been extensively explored recently (Wang
et al., 2021)(J
¨
ager and Krems, 2023).We are inter-
ested in Quantum Embedding Kernels (QKEs), which
use quantum circuits to embed data points into a
high-dimensional Hilbert space. The overlap between
these quantum states is then used to compute the in-
ner product between data points in this feature space.
Typically, these kernels are parameterized, making
them a form of VQCs. The parameters of these cir-
cuits are optimized based on Kernel Target Alignment
(KTA), which serves as a metric to align the kernel
with the target task (which we will consider to be bi-
nary classification) (Hubregtsen et al., 2022). Once
the kernel is optimized, it is fed into an SVM to de-
termine the optimal decision boundary for the classi-
fication task at hand. This integration leverages the
strengths of both quantum and classical computing,
aiming to enhance classification performance due to
the expressive power of quantum embeddings com-
bined with the robust framework of SVMs.
However, the method scales poorly. For instance,
using the KTA as cost function requires calculat-
ing the kernel matrix at every single training step, a
process that scales quadratically O(N
2
) with train-
ing dataset size N. To alleviate this, both the origi-
nal paper (Hubregtsen et al., 2022) and a following
one (Sahin et al., 2024) propose using only a subsam-
ple of size D ≪ N of the training dataset to compute
the KTA at each epoch, effectively turning the com-
putation, which becomes O(D
2
), independent of N.
Moreover, another paper (Tscharke et al., 2024) pro-
poses using a clustering algorithm to find centroids
of the classes and then computing the kernel matrix
with respect to these centroids at each training step,
bringing the complexity of the computation to O(N).
So, these methods are able to decrease the complex-
ity of training from scaling quadratically with N to
either scaling only linearly or even being completely
independent of N. Nonetheless, the end goal is for
the optimized kernel matrix, which contains the pair-
wise inner-products between all training data points,
to be fed into the SVM. Thus, at least for this final
computation, these methods still require O(N
2
) com-
putations. This is especially important if we consider
that each pairwise inner-product requires a quantum
circuit execution.
In this paper, we adapt a low-rank matrix approx-
imation known as the Nystr
¨
om Approximation, which
is commonly used in classical kernel methods, to ad-
dress this scalability issue. This approach reduces
the complexity of computing the kernel matrix from
O(N
2
) to O(NM
2
), where M ≪ N is an hyperparam-
eter that determines the quality of the approximation.
The Nystr
¨
om Approximation is applied for comput-
ing the optimized kernel matrix that is fed into the
SVM, resulting in a classification pipeline that scales
linearly with the training dataset size N in all steps.
Consequently, our method facilitates the efficient ap-
plication of quantum kernel methods to industrially-
relevant problems.
The contributions of this paper are organized as
follows:
• Adapted the Nystr
¨
om Approximation to Quantum
Embedding Kernels
• Empirically verified (on several synthetic
datasets) that, in a noiseless setting, the Nystr
¨
om
Approximation method allows for quantum
kernels with reduced quantum circuit executions
at a small cost in the accuracy of the resulting
SVM.
• Empirically tested the performance of the devel-
oped method under both coherent and incoherent
noise.
2 KERNEL METHODS AND
SUPPORT VECTOR MACHINES
Kernel methods can be used for different tasks, from
dimensionality reduction (Sch
¨
olkopf et al., 1997) to
regression (Drucker et al., 1996). We will focus on bi-
nary classification using SVMs, which are linear clas-
sifiers (Hearst et al., 1998). Nonetheless, they can be
used in non-linear classification problems due to the
kernel trick, which implicitly maps the data into high-
dimensional feature spaces where linear classification
is possible. In this section we will go through this
pipeline for classification, starting with kernel meth-
ods and ending with SVMs.
2.1 Kernel Methods
Consider a dataset X = {(x
i
, y
i
)}
n
i=1
, where x
i
∈ R
d
,
with d being the number of features of x
i
. A kernel
method involves defining a feature map φ : R
d
→ H ,
where H is a high-dimensional Hilbert space. The
kernel function can then be defined as:
k(x
i
, x
j
) = ⟨φ(x
i
), φ(x
j
)⟩
H
(1)
Put in words, the kernel function computes the
inner product between the inputs x
i
and x
j
in some
high-dimensional feature space H . Given the kernel
function k, the kernel matrix K is a symmetric matrix
that contains the pairwise evaluations of k over all the
points in the training dataset X:
QAIO 2025 - Workshop on Quantum Artificial Intelligence and Optimization 2025
764

K
i j
= K(x
i
, x
j
) = ⟨φ(x
i
), φ(x
j
)⟩
H
(2)
Typically, explicitly calculating the coordinates of
the input points in the high-dimensional feature space,
a task that may be computationally expensive, is not
needed. Instead, the kernel trick allows one to effi-
ciently compute these inner products without needing
to explicitly calculate the feature map φ.
For example, consider the Radial Basis Function
(RBF) kernel, defined as:
k
RBF
(x, y) = exp
−γ∥x − y∥
2
(3)
where γ is an hyperparameter that the user defines.
This kernel computes the inner product between the
data points x and y in an infinite-dimensional feature
space given by the feature map ϕ
RBF
. However, due to
the kernel trick, one only needs to compute the kernel
k
RBF
and not the feature map ϕ
RBF
, saving computa-
tional resources.
2.2 Support Vector Machines
An SVM aims to find the optimal hyperplane that sep-
arates data points of two different classes with the
maximum margin. This margin is the distance be-
tween the hyperplane and the nearest data points from
either class, known as support vectors (Hearst et al.,
1998). SVMs are inherently linear classifiers; they
work effectively when the dataset is linearly separa-
ble by a hyperplane.
However, the kernel trick extends SVMs to non-
linear datasets. By using a kernel function, the data
is implicitly mapped into a high-dimensional feature
space where linear separation is feasible. Thus, with
the aid of the kernel trick, SVMs can classify non-
linear datasets.
Given a kernel matrix K containing the pairwise
inner-products between all training points x ∈ X, the
optimization problem the SVM solves is formulated
as:
min
α
1
2
n
∑
i=1
n
∑
j=1
α
i
α
j
y
i
y
j
K(x
i
, x
j
) −
n
∑
i=1
α
i
(4)
where α
i
are the Lagrange multipliers. An SVM
solves a quadratic problem and thus is guaranteed to
converge to the global minimum.
The decision function is given by:
f (x) = sign
n
∑
i=1
α
i
y
i
K(x
i
, x) + b
!
(5)
Here, b is the bias term, often determined using
the support vectors. However, as shown in Equation
4, training an SVM on a dataset of size N requires
Figure 1: One layer of the quantum ansatz used through-
out this work. As an example, this layer in particular con-
tains 4 qubits. The ansatz contains input scaling parameters
λ and variational parameters θ. The data encoding gates
(blue-colored) are repeated in every single layer - data re-
uploading.
computing N
2
pairwise inner products to construct the
kernel matrix K. This implies that the runtime scales
at least quadratically with the training dataset size N.
Moreover, even after obtaining the kernel matrix K,
the SVM must solve a quadratic optimization prob-
lem. Depending on the data structure and the algo-
rithm used, this process may scale with N
2
or even N
3
.
Consequently, SVMs are typically applied to small-
scale problems with moderate dataset sizes.
3 QUANTUM EMBEDDING
KERNELS
The first step to solve a classical problem using a
quantum computer is to encode the classical data into
a quantum state that can be processed by quantum op-
erations. Given a data point x, one can define a quan-
tum circuit U(x) to generate said quantum state:
|
ϕ(x)
⟩
= U(x)
|
0
⟩
(6)
This corresponds to embedding the data in a high-
dimensional Hilbert space. Referring back to Equa-
tion 1, a kernel function is defined as the inner prod-
uct between two data points in a high-dimensional
Hilbert space. In the context of quantum computing,
this translates to calculating the fidelity between the
quantum states generated by encoding the two data
points. The fidelity, which measures the similarity be-
tween these quantum states, is given by:
k(x
i
, x
j
) =
⟨ϕ(x
i
)|ϕ(x
j
)⟩
2
(7)
There are several ways to calculate the fidelity be-
tween two quantum states. In particular, assuming
that ϕ(x) and ϕ(y) are pure quantum states (that is,
Trace(ρ
2
) = 1, where ρ =
⟨
ϕ
|
ϕ
⟩
), then the fidelity can
be calculated using the adjoint of the quantum circuit
(Hubregtsen et al., 2022):
K(x, y) =
|
⟨ϕ(x)|ϕ(y)⟩
|
2
=
⟨0|U
†
(y)U(x)|0⟩
2
(8)
Quantum-Efficient Kernel Target Alignment
765
(a) . (b) .
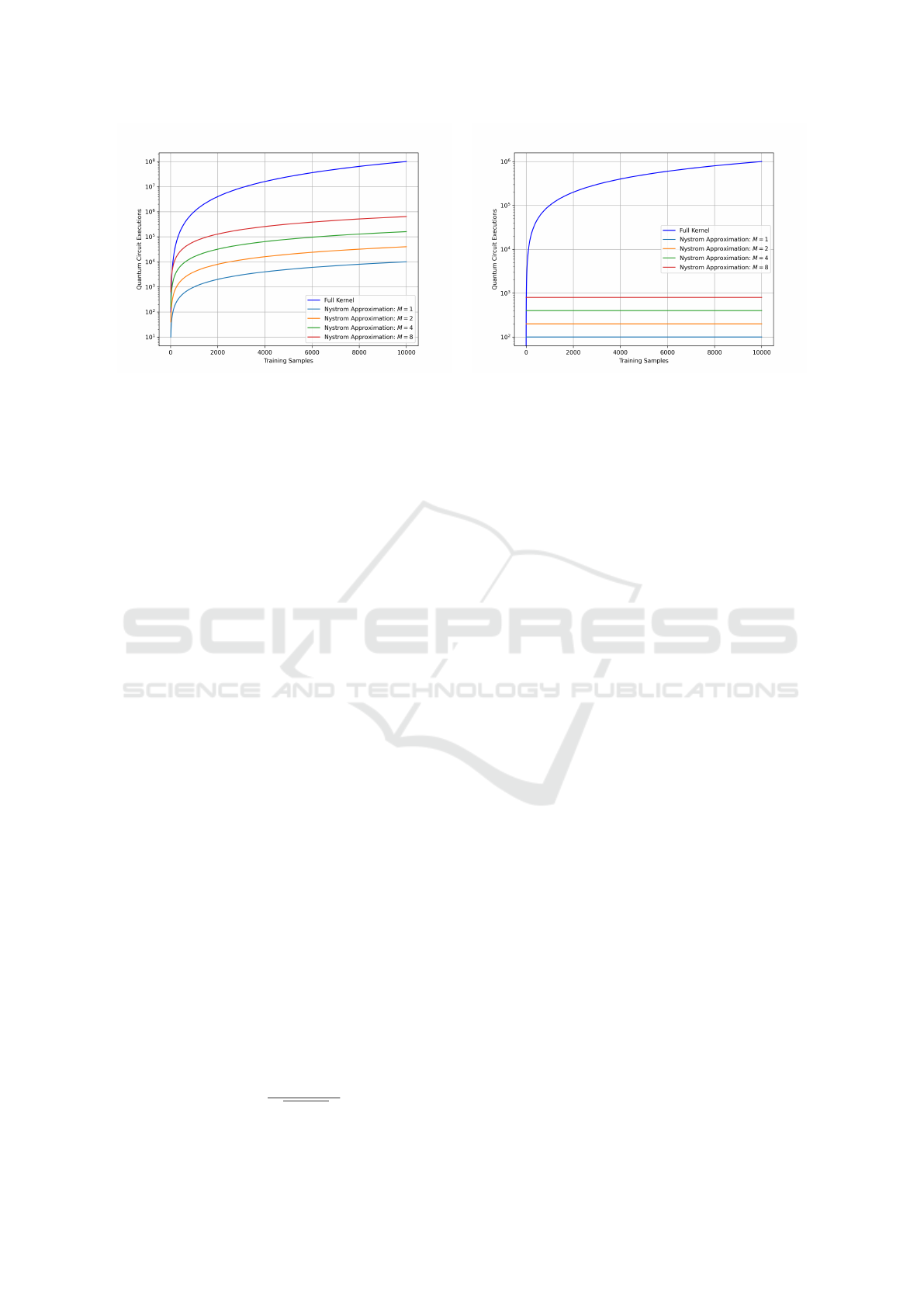
Figure 2: Quantum circuit executions required for computing the training kernel matrix (Fig.2a) and testing kernel matrix
(Fig.2b) as a function of the training dataset size N for the standard approach and the Nystr
¨
om approximation with different
Ms.
This is equivalent to the probability of observing
|
0
⟩
when measuring U
T
(y)U(x)
|
0
⟩
in the computa-
tional basis.
We have established a definition for quantum ker-
nels. Nevertheless, the choice of which quantum ker-
nel to use remains unclear. In Noisy-Intermediate
Scale Quantum (NISQ) devices, a popular approach
is to make the circuit trainable, define a cost function,
and optimize the parameters using a classical opti-
mizer. These methods are known as Variational Quan-
tum Algorithms. In our specific case, a quantum ker-
nel can be made trainable by making the quantum cir-
cuit U (x) also depend on these classical parameters,
becoming U(x, θ). Thus, the kernel function from Eq.
8 becomes:
K
θ
(x, y) =
⟨0|U
†
(y,θ)U(x, θ)|0⟩
2
(9)
Then, the question becomes what cost function to
use. Following paper (Hubregtsen et al., 2022), we
use Kernel-Target Alignment (KTA) as our cost func-
tion, which predicts the alignment between the quan-
tum kernel K
θ
and the labels of the training data. We
start by defining an ideal kernel using the labels of the
training data:
k
y
(x
i
, x
j
) = y
i
y
j
(10)
Then, assuming y ∈ {−1, 1}, k
y
will be 1 if both
labels belong to the same class and −1 otherwise.
This ideal kernel matrix can be defined as:
K
y
= y
T
y (11)
Then, the KTA can be defined as:
KTA =
y
T
Ky
p
Tr(K
2
)N
(12)
In this context, N represents the total number of
training samples in the dataset. Since we want to min-
imize a cost function and maximize KTA, we will in-
clude a negative sign in this equation during training.
4 NYSTR
¨
OM APPROXIMATION
To reduce the complexity of computing the ker-
nel matrix, one can use the Nystr
¨
om approxima-
tion (Drineas et al., 2005). It works as follows.
The first step is to select a subset of M ≪ N data
points from the original training dataset N, referred
to as landmarks. In this work, we will always
select the landmarks randomly, but they can also
be selected according to more sophisticated strate-
gies, such as K-means clustering. Then, compute
the matrix K
MM
of shape (M, M) using the selected
subset where, given landmarks {m
1
, m
2
, ..., m
M
},
K
MM
(i, j) = ⟨φ(m
i
), φ(m
j
)⟩
H
. Then, compute the
cross-kernel matrix K
NM
of shape (N, M) between the
M landmarks and the N training data points. Finally,
the Nystr
¨
om approximation of the kernel matrix K is
given by:
˜
K ≈ K
NM
K
−1
MM
K
T
NM
(13)
If we disregard the computational cost of invert-
ing K
MM
(as this step is performed classically), the
Nystr
¨
om approximation reduces the computational
cost of calculating the kernel matrix from O(N
2
) to
O(NM
2
). Consequently, the number of quantum cir-
cuit executions required to compute the matrix now
scales linearly with N, significantly enhancing effi-
ciency in the quantum context.
This method can also be used to reduce the com-
putational cost of generating the kernel matrix to clas-
QAIO 2025 - Workshop on Quantum Artificial Intelligence and Optimization 2025
766
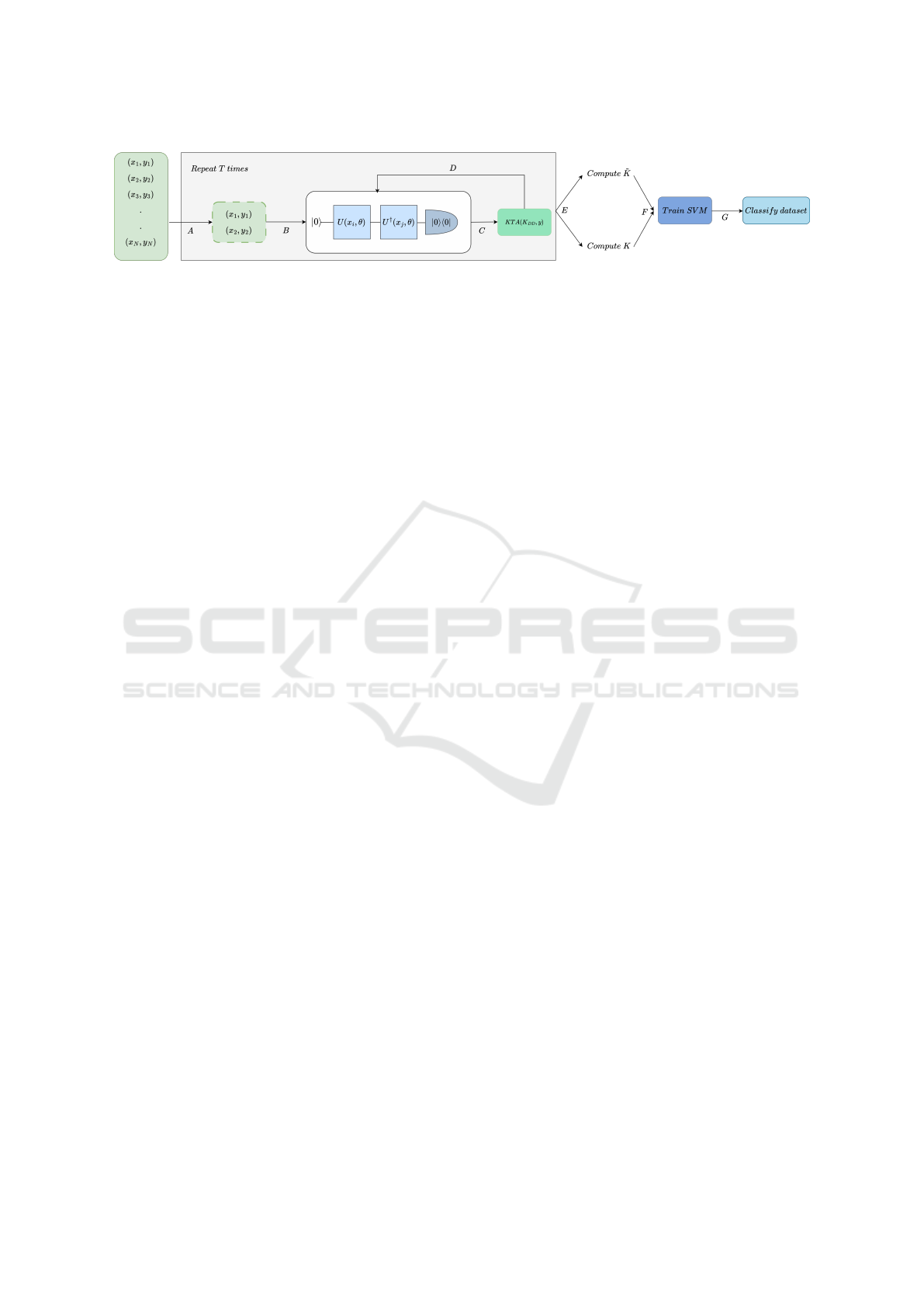
Figure 3: Given a training dataset of size N, for T iterations, do: A) sample a random mini-batch of data points of size D. B)
Calculate the overlap between the embedded quantum states. C) Once all the pairs have been processed, fill the kernel matrix
K
DD
and compute the cost function (KTA). D) Update the parameters θ of the quantum circuit. Finally, after the T training
iterations, E) Compute the approximated training kernel matrix
˜
K using either the Nystr
¨
om approximation method or the full
training kernel matrix K using the standard approach. F) Use the training kernel matrix to fit an SVM to the dataset and G)
Classify the training dataset.
sify a test dataset. The end goal in classification tasks
is to test the model on unseen data points. With an
SVM, this requires generating a testing kernel ma-
trix of shape (P, N), where P is the size of the test-
ing dataset, leading to a computational complexity of
O(PN). However, using the Nystr
¨
om approximation,
one can reduce this computational cost to O(PM),
making it independent of the training dataset size N.
This is accomplished by computing:
˜
K
test
≈ K
T
PM
K
−1
MM
K
NM
(14)
Here, K
PM
is the cross-kernel matrix between the
M landmarks and the P test data points, and K
MN
is
the previously computed cross-kernel matrix between
the M landmarks and the N training data points. For
a comparison in terms of the number of quantum cir-
cuit executions required to construct the training and
testing kernel matrices using either the Nystr
¨
om ap-
proximation or the standard approach, see Fig.2
5 METHOD
In this work, we test the Nystr
¨
om approximation
method for generating the kernel matrix that is fed
into the SVM in the context of quantum embedding
kernels. We start with a training dataset with N data
points and an ansatz for the VQC, see Fig. 1, ran-
domly initializing the free parameters. The quantum
circuit is then trained over T training iterations using
the KTA as the cost function.
However, calculating the kernel matrix K for ev-
ery training iteration is computationally expensive.
To mitigate this, we adopt a strategy similar to that
used in (Hubregtsen et al., 2022; Sahin et al., 2024).
At each iteration, we randomly sample a mini-batch
of D training points, construct their kernel matrix
˜
K
DD
, calculate the KTA for this mini-kernel, and up-
date the parameters using a classical optimizer. Con-
sequently, the number of quantum circuit executions
per training iteration becomes independent of the size
of the training dataset N and instead scales quadrati-
cally with D. Since, as we will see, D can be chosen
such that D ≪ N, this method significantly reduces
the number of quantum circuit executions required
per training iteration.
After completing the T training iterations and op-
timizing the quantum kernel for the task at hand,
we compute the kernel matrix for the entire training
dataset N using the Nystr
¨
om approximation method.
This is our main contribution in this work. Using this
approximation allows us to build the kernel matrix
that is fed into the SVM at the end of training using
only O(NM
2
) (where M is the number of landmarks),
quantum circuit executions, instead of the O(N
2
) exe-
cutions that both (Hubregtsen et al., 2022) and (Sahin
et al., 2024) require. To our knowledge, this is the first
pipeline in which the number of quantum circuit ex-
ecutions scales linearly with the training dataset size
N in all steps. Specifically, the training process has a
complexity of O(D
2
), and the computation of the final
kernel matrix of O(NM
2
) (note that this complexity
takes into account only the number of quantum cir-
cuit executions). The entire pipeline can be seen in
Fig.3.
Although not explicitly shown in the figure, the
Nystr
¨
om approximation method saves quantum cir-
cuit executions in comparison with the standard ap-
proach during inference as well, when classifying a
testing dataset. The standard approach has a (quan-
tum) complexity of O(NP), with P being the size of
the testing dataset, while the Nystr
¨
om approximation
has a complexity of O(PM).
Lastly, we wish to explain in higher depth the dif-
ferences between the method from (Sahin et al., 2024)
and the Nystr
¨
om approximation method we propose
here. In short, they are different methods with dif-
ferent applications. The method from (Sahin et al.,
2024) allows us to use KTA as the cost function with-
out computing a full kernel matrix at each training
step. Instead, it computes a mini-kernel matrix K
DD
,
reducing quantum circuit executions from O(N
2
) to
Quantum-Efficient Kernel Target Alignment
767
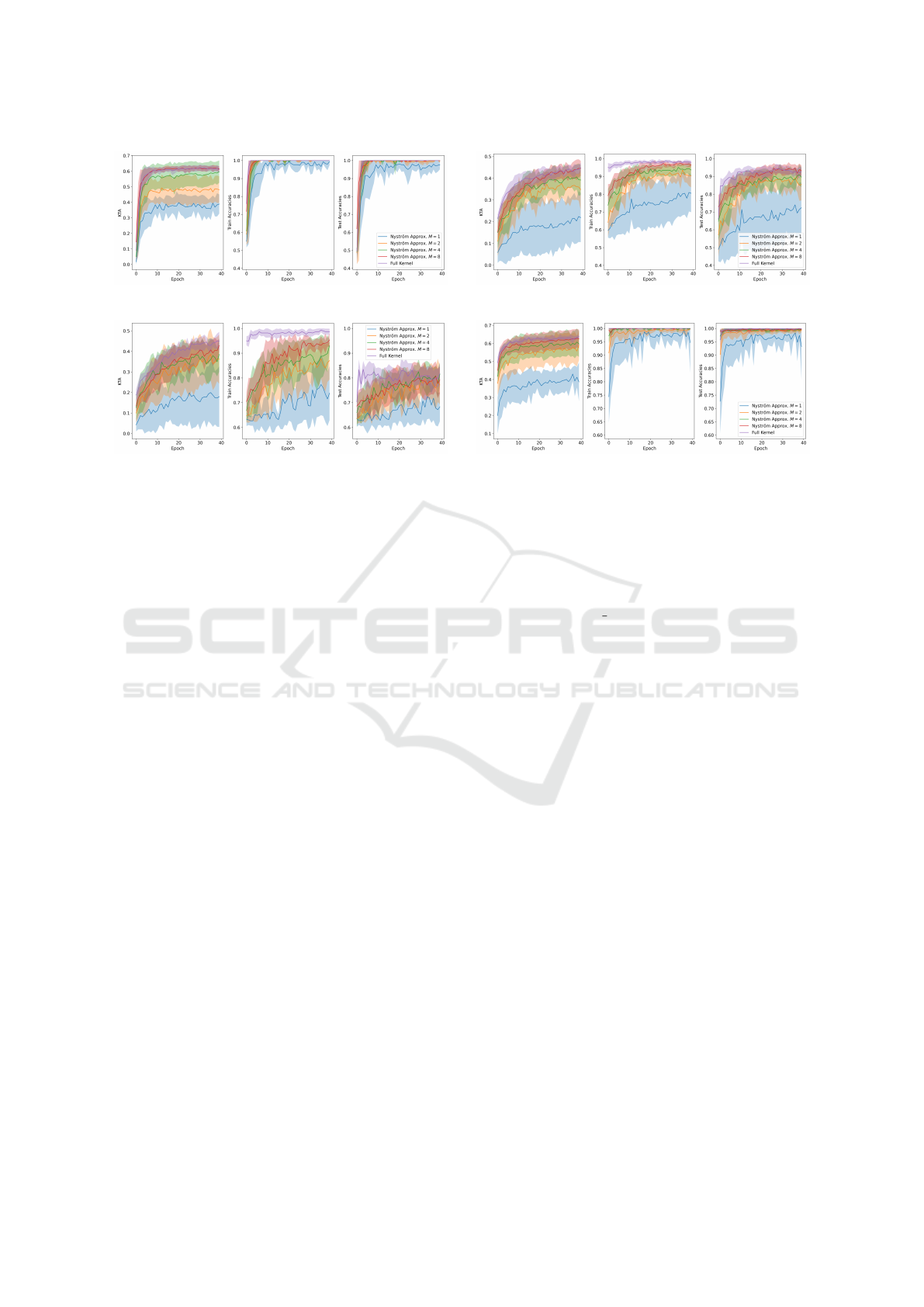
(a) . (b) .
(c) . (d) .
Figure 4: Performance of the Standard approach VS the Nystr
¨
om approximation with different Ms throughout training mea-
sured through KTA, training and testing accuracy in the checkers (Fig. 4a), corners (Fig. 4b), donuts (Fig. 4c) and spirals
(Fig. 4d) datasets.
O(D
2
), where D ≪ N. This is essentially a type of
stochastic gradient descent, where KTA is approxi-
mated by using a subset of data points.
After training, the SVM requires the full opti-
mized kernel matrix to classify the training dataset.
Therefore, we cannot use the previous method here,
as the SVM needs the complete matrix (O(N
2
) quan-
tum circuit executions) rather than a mini-version
based on a subsample. The Nystr
¨
om Approximation
method addresses this need by computing an approxi-
mated full-kernel matrix
˜
K while reducing the number
of quantum circuit executions to O(NM
2
). Thus, us-
ing both methods —(Sahin et al., 2024) during train-
ing and the Nystr
¨
om Approximation for generating
the kernel matrices once training is done —results in
a pipeline that scales linearly with the training dataset
size in all steps.
We can also apply the Nystr
¨
om Approximation
during the T training iterations instead of the method
from (Sahin et al., 2024) by using the approximated
kernel matrix
˜
K to compute KTA. However, this does
not reduce quantum circuit executions and, based on
limited experiments, did not yield significantly better
results. Therefore, in this work, we use the method
from (Sahin et al., 2024) during training.
6 NUMERICAL RESULTS
In this section, we empirically compare the Nystr
¨
om
approximation method and the standard method on
a set of simple binary 2D classification tasks. The
datasets selected include the checkers and donuts
datasets from (Hubregtsen et al., 2022), a manually
created corners dataset, and the spirals dataset gener-
ated using the make moons method from scikit-learn.
For a more detailed explanation, please refer to Ap-
pendix 8.
To compare the methods, we will use three met-
rics: the KTA between the current quantum kernel
and the ideal kernel for the entire training dataset, the
training accuracy, and the testing accuracy. Note that
calculating the first two metrics requires computing
the training kernel matrix, while the last metric re-
quires computing the testing kernel matrix. This is
where the standard approach and the Nystr
¨
om approx-
imation differ, as previously explained. Due to the in-
herent randomness of parameter initialization, results
are averaged over 10 seeds and plotted alongside the
standard deviation.
For the experiments, the input scaling weights are
initialized as 1s, ensuring they initially have no effect
on the output of the quantum kernel, and the varia-
tional weights sampled uniformly between −π and π.
The methods were implemented using PyTorch and
PennyLane and all results obtained using state-vector
simulators. The parameters were optimized using the
ADAM optimizer with a learning rate of 0.1 and the
ansatz had a constant depth of 5 layers. Moreover,
during the training iterations, we always used a mini-
batch size D = 8.
QAIO 2025 - Workshop on Quantum Artificial Intelligence and Optimization 2025
768
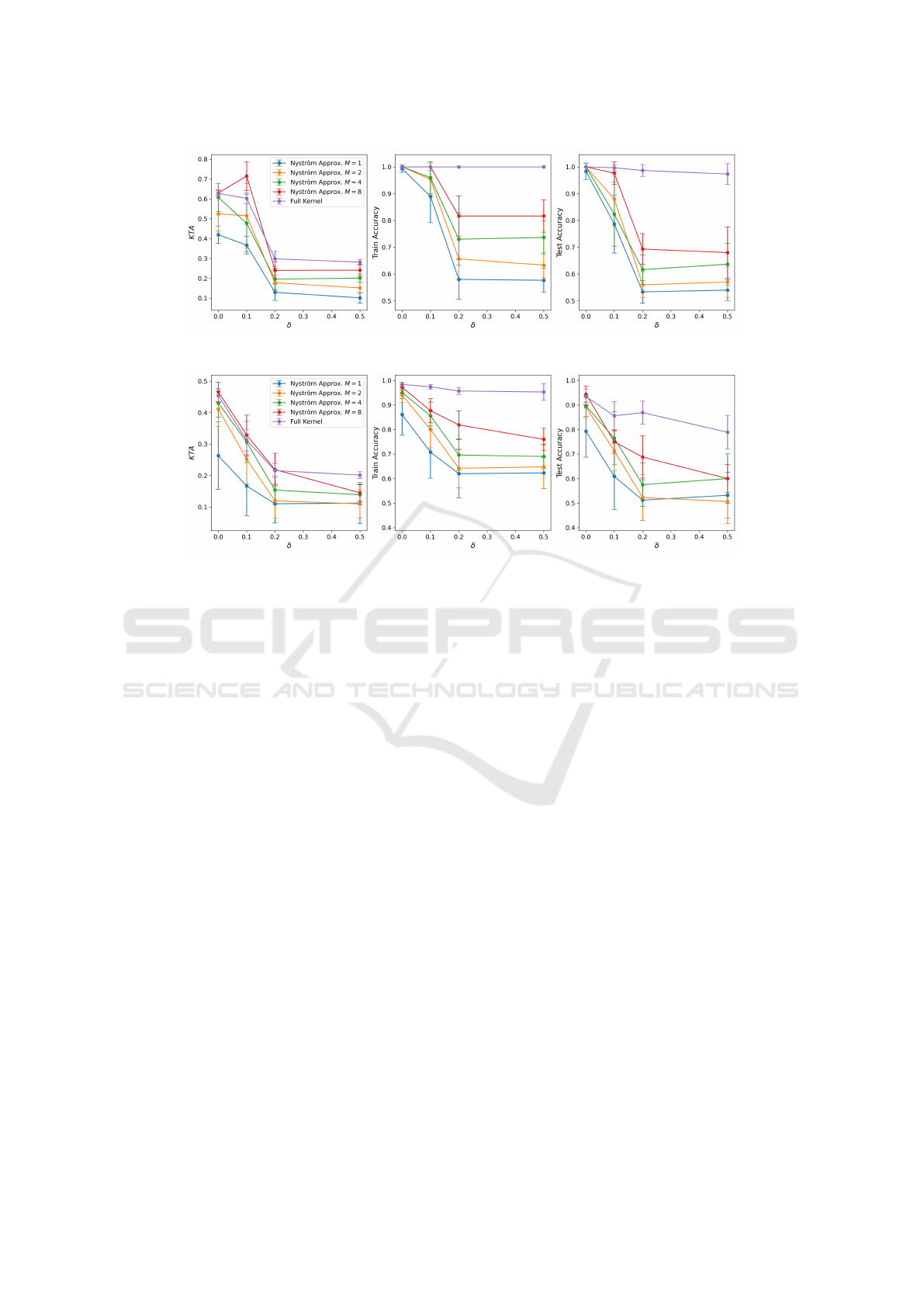
(a) .
(b) .
Figure 5: Performance of the Standard approach VS the Nystr
¨
om approximation with different Ms for an increasing strength
of coherent noise σ in the checkers (Fig.5a) and corners dataset (Fig.5b).
6.1 Method Comparison on Noiseless
Simulator
We will start by analyzing both methods in ideal con-
ditions, that is, in the absence of noise.
The performance comparison between the stan-
dard approach and the Nystr
¨
om approximation on
four benchmark datasets is illustrated in Fig.4.
As observed in the KTA graphs, the standard ap-
proach consistently achieves the highest KTA across
all four datasets. Moreover, a lower value of M typ-
ically results in a reduced KTA for the Nystr
¨
om ap-
proximation method. This is expected, as lower val-
ues of M lead to less accurate approximations of the
training and testing kernel matrices. However, this
lower KTA does not necessarily imply reduced accu-
racies, as evidenced by the training and testing results.
In some datasets, the Nystr
¨
om method achieves both
training and testing accuracies of 100%, despite the
lower KTA.
These findings suggest an important considera-
tion: if one is willing to potentially sacrifice a slight
amount of accuracy for a significantly reduced num-
ber of quantum circuit executions, the Nystr
¨
om Ap-
proximation should be considered. Moreover, it’s also
clear that the training approach from (Sahin et al.,
2024) of using a mini-batch of size D every train-
ing step works, since in all datasets both accuracy and
KTA increased throughout training.
However, some nuances should be discussed. It’s
not clear precisely how both M and D are related to
the datasets’ complexity and/or size. We used sev-
eral training datasets with sizes ranging from 30 to
100 training data points and observed that, for all of
them, D = 8 and M ∈ {2, 4, 8} lead to considerably
high accuracies, achieving near or perfect classifica-
tion on most of the datasets. However, for larger
and more complex datasets, it may be necessary to
increase both the values of D and M. It may also
be helpful to select the D data points and M land-
marks not randomly but instead based on more so-
phisticated techniques. For instance, the M landmarks
for the Nystr
¨
om approximation technique can be se-
lected using k-means clustering, ensuring that they
are representative of the entire dataset, effectively bet-
ter approximating the kernel matrix. However, test-
ing such techniques is out of the scope of this work,
which intends to simply demonstrate for the first time
a quantum-kernel pipeline that depends only linearly
on the training dataset size. In short, we do not claim
that the values of M and D used in this work and the
technique we used to select them (uniformly random
sampling) are guaranteed to provide good results for
any dataset. In fact, this pipeline should be highly
Quantum-Efficient Kernel Target Alignment
769

dependent upon the dataset and, consequently, these
hyperparameters should be carefully chosen.
These results are in ideal conditions, if one had
access to a perfect quantum computer. In the next
subsections we will look at how the methods compare
under different types of noise.
6.2 Method Comparison Under
Coherent Noise
Coherent noise are errors that preserve the unitary
evolution of the quantum circuit but that still affect its
output (Cai et al., 2020). In (Skolik et al., 2023), co-
herent noise was modeled as under or over-rotations
of the parametrized quantum gates, which can be seen
as a miscalibration of the quantum gates. We will fol-
low a similar approach. The variational quantum pa-
rameters θ will be changed according to θ → θ + δθ,
where δθ are sampled according to a Gaussian distri-
bution of mean 0 and variance σ
2
. The perturbations
δθ are re-sampled if a new observable is being mea-
sured or if the circuit is being evaluated with a differ-
ent set of parameters (Skolik et al., 2023).
The performance of both the standard approach
and the Nystr
¨
om approximation under increasing
strength of coherent noise σ can be seen in Fig. 5.
Firstly, it is evident that the standard approach
demonstrates better resistance to coherent noise. For
both datasets, although the KTA decreases signifi-
cantly as σ increases, the training and testing accu-
racy only slightly decline for the corners dataset. In
contrast, the Nystr
¨
om approximation method shows
a sharp decrease in KTA, training, and testing ac-
curacy when σ > 0.2. Furthermore, as observed in
the noiseless setting, lower values of M tend to per-
form worse. These results are unsurprising, as the
Nystr
¨
om method approximates the kernel matrix us-
ing only a limited number of quantum circuit execu-
tions. If these executions are noisy, it is expected that
the resulting approximation will be even less accu-
rate. In summary, the approximation and the noise
compound, whereas the standard approach can toler-
ate more noise.
6.3 Method Comparison Under
Depolarizing Noise
In this section, we aim to analyze the performance
of both methods under a different type of noise: De-
polarizing Noise. This noise affects a quantum state
by replacing it with a mixed state with probability p,
or leaving it unchanged otherwise. It’s important to
highlight that, in the presence of device noise, the ad-
joint operation required to compute the quantum ker-
nel function may not be feasible. However, as shown
in (Hubregtsen et al., 2022), it is indeed possible in
the case of depolarizing noise.
In Pennylane, depolarizing noise can be imple-
mented using the Depolarizing Channel operation.
We model this type of noise by introducing single-
qubit depolarizing channels after each quantum gate.
The performance of both methods under depolar-
izing noise as a function of the probability p can be
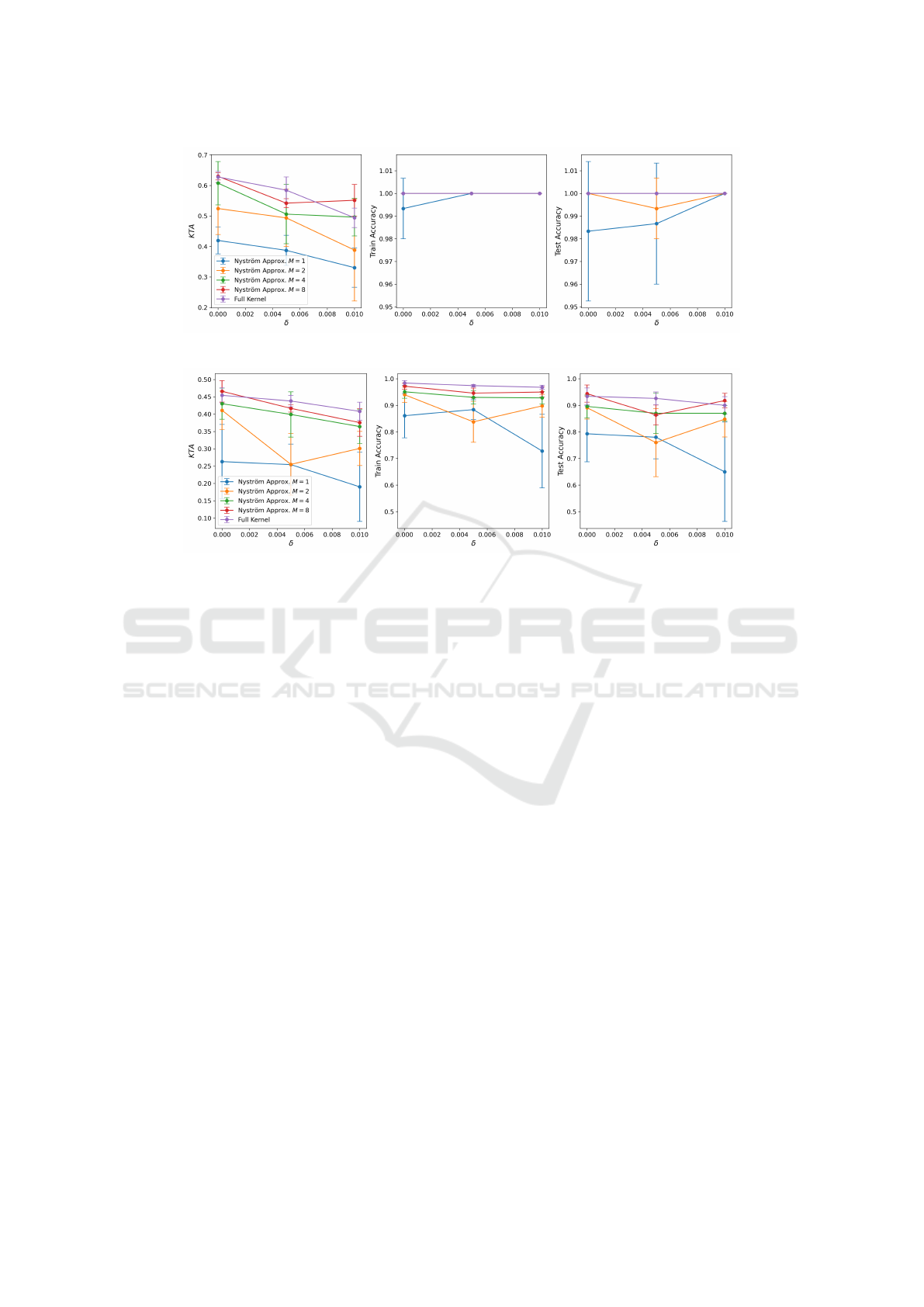
seen in Fig.6.
Here too, similar results can be observed. The
standard approach is more robust against depolariz-
ing noise when compared with the Nystr
¨
om method.
Nonetheless, both methods are able to effectively
reach high training and testing accuracies. In particu-
lar, if one uses the Nystr
¨
om method with a larger M,
then the results are at least comparable to the stan-
dard approach, while still requiring substantially less
quantum circuit executions.
7 CONCLUSION
In this work we adapted a low-rank matrix approx-
imation method named Nystr
¨
om approximation to
quantum embedding kernels, effectively reducing the
number of quantum circuit executions required to
construct the training and testing kernel matrices. In
doing so, we also introduce the first quantum ker-
nel pipeline that has an end-to-end quantum complex-
ity that depends only linearly on the training dataset
size N. Other works had already introduced a linear
complexity during the training stage, but not during
the training matrix creation. Furthermore, we show
that, in the noiseless scenario, an SVM trained from a
quantum kernel using the Nystr
¨
om approximation has
a performance comparable to that of an SVM trained
using the standard quantum kernel approach, without
any approximation. Finally, we compare both meth-
ods in the presence of coherent and depolarizing noise
and empirically demonstrate that both methods per-
form well under realistic levels of noise.
CODE AVAILABILITY
The source code for replicating the results is available
on Github at https://github.com/RodrigoCoelho7/
qekta.
QAIO 2025 - Workshop on Quantum Artificial Intelligence and Optimization 2025
770
(a) .
(b) .
Figure 6: Performance of the Standard approach VS the Nystr
¨
om approximation with different Ms for an increasing proba-
bility of depolarizing noise p in the checkers (Fig.6a) and corners dataset (Fig.6b).
ACKNOWLEDGEMENTS
The research is part of the Munich Quantum Valley,
which is supported by the Bavarian state government
with funds from the Hightech Agenda Bayern Plus.
REFERENCES
Abbas, A., Sutter, D., Zoufal, C., Lucchi, A., Figalli, A.,
and Woerner, S. (2021). The power of quantum neural
networks. Nature Computational Science, 1(6):403–
409.
Cai, Z., Xu, X., and Benjamin, S. C. (2020). Mitigating
coherent noise using pauli conjugation. npj Quantum
Information, 6(1):17.
Cerezo, M., Arrasmith, A., Babbush, R., Benjamin, S. C.,
Endo, S., Fujii, K., McClean, J. R., Mitarai, K., Yuan,
X., Cincio, L., et al. (2021). Variational quantum al-
gorithms. Nature Reviews Physics, 3(9):625–644.
Dhillon, I. S., Guan, Y., and Kulis, B. (2004). Kernel k-
means: spectral clustering and normalized cuts. In
Proceedings of the tenth ACM SIGKDD international
conference on Knowledge discovery and data mining,
pages 551–556.
Drineas, P., Mahoney, M. W., and Cristianini, N. (2005). On
the nystr
¨
om method for approximating a gram matrix
for improved kernel-based learning. journal of ma-
chine learning research, 6(12).
Drucker, H., Burges, C. J., Kaufman, L., Smola, A., and
Vapnik, V. (1996). Support vector regression ma-
chines. Advances in neural information processing
systems, 9.
Farhi, E., Goldstone, J., and Gutmann, S. (2014). A
quantum approximate optimization algorithm. arXiv
preprint arXiv:1411.4028.
Grover, L. K. (1996). A fast quantum mechanical algorithm
for database search. In Proceedings of the twenty-
eighth annual ACM symposium on Theory of comput-
ing, pages 212–219.
Hearst, M. A., Dumais, S. T., Osuna, E., Platt, J., and
Scholkopf, B. (1998). Support vector machines. IEEE
Intelligent Systems and their applications, 13(4):18–
28.
Hubregtsen, T., Wierichs, D., Gil-Fuster, E., Derks, P.-J. H.,
Faehrmann, P. K., and Meyer, J. J. (2022). Train-
ing quantum embedding kernels on near-term quan-
tum computers. Physical Review A, 106(4):042431.
J
¨
ager, J. and Krems, R. V. (2023). Universal expressive-
ness of variational quantum classifiers and quantum
kernels for support vector machines. Nature Commu-
nications, 14(1):576.
Kandala, A., Mezzacapo, A., Temme, K., Takita, M.,
Brink, M., Chow, J. M., and Gambetta, J. M. (2017).
Hardware-efficient variational quantum eigensolver
Quantum-Efficient Kernel Target Alignment
771

for small molecules and quantum magnets. nature,
549(7671):242–246.
Preskill, J. (2018). Quantum computing in the nisq era and
beyond. Quantum, 2:79.
Sahin, M. E., Symons, B. C., Pati, P., Minhas, F., Mil-
lar, D., Gabrani, M., Mensa, S., and Robertus, J. L.
(2024). Efficient parameter optimisation for quantum
kernel alignment: A sub-sampling approach in varia-
tional training. Quantum, 8:1502.
Sch
¨
olkopf, B., Smola, A., and M
¨
uller, K.-R. (1997). Kernel
principal component analysis. In International con-
ference on artificial neural networks, pages 583–588.
Springer.
Schuld, M. (2018). Supervised learning with quantum com-
puters. Springer.
Shor, P. W. (1999). Polynomial-time algorithms for prime
factorization and discrete logarithms on a quantum
computer. SIAM review, 41(2):303–332.
Skolik, A., Jerbi, S., and Dunjko, V. (2022). Quantum
agents in the gym: a variational quantum algorithm
for deep q-learning. Quantum, 6:720.
Skolik, A., Mangini, S., B
¨
ack, T., Macchiavello, C., and
Dunjko, V. (2023). Robustness of quantum reinforce-
ment learning under hardware errors. EPJ Quantum
Technology, 10(1):1–43.
Tscharke, K., Issel, S., and Debus, P. (2024). Quack:
Quantum aligned centroid kernel. arXiv preprint
arXiv:2405.00304.
Wang, X., Du, Y., Luo, Y., and Tao, D. (2021). Towards un-
derstanding the power of quantum kernels in the nisq
era. Quantum, 5:531.
APPENDIX
The four binary 2D datasets used in this work for
benchmarking the models can be seen on Fig.7. A
specification of the datasets and how to create them:
• Checker from (Hubregtsen et al., 2022). This
dataset consists of 30 training and 30 testing data
points. Used on both noiseless and noisy numeri-
cal results. The reader is referred to (Hubregtsen
et al., 2022) for more details on how to create it.
• Donuts from (Hubregtsen et al., 2022). This
dataset consists of 60 training and 60 testing data
points. Used on noiseless numerical results. The
reader is referred to (Hubregtsen et al., 2022) for
more details on how to create it.
• Spirals from scipy’s make moons method. This
dataset consists of 100 training and 100 testing
data points. Used on noiseless numerical results.
• Corners. This dataset consists of 100 training and
100 testing data points. To generate the dataset,
we first define a square of size 2 (both x and y-
axis are between −1 and 1) and randomly sample
a set of points within this region. Each point is
then classified based on its location relative to four
quarter circles, each centered at one of the corners
of the square. Specifically, if a point falls within
any of these quarter circles, it is labeled as −1;
otherwise, it is labeled as 1. Used on noiseless
and noisy
(a) Checkers Dataset. (b) Donuts Dataset.
(c) Spirals Dataset. (d) Corners Dataset.
Figure 7: Red and blue represent the two classes. Cir-
cles/Circumferences represent training/testing data points.
QAIO 2025 - Workshop on Quantum Artificial Intelligence and Optimization 2025
772
