
Physiology-Guided Blood Glucose Predictive Model Using Minimal
Blood Glucose Dynamics
Sarala Ghimire
1
, Turgay Celik
2
, Martin Gerdes
1
and Christian W. Omlin
2
1
Department of Information and Communication Technologies, Centre for e-Health, University of Agder,
Grimstad, Norway
2
Department of Information and Communication Technologies, Centre for Artificial Intelligence Research (CAIR),
University of Agder, Grimstad, Norway
Keywords: Blood Glucose, Data-Driven, Hybrid, Mathematical Model, Prediction, Physiology.
Abstract: Modelling blood glucose and insulin dynamics using mathematical equations requires a deep understanding
of individual physiology and relying on numerous predefined parameters necessitating extensive clinical and
personal data, making direct use of these models for blood glucose prediction computationally intensive and
inaccurate. Though data-driven models are more efficient and require no individual physiology, they produce
predictions that are inconsistent with known glucose-insulin interactions. Thus, this study aims to investigate
the potentiality of physiological models integrated with data-driven approach for predicting blood glucose
level. It intends to extract simple physiological dynamics of blood glucose kinetics and incorporate them into
a data-driven model, with less reliance on detailed individual data. The result demonstrated that the model
integrating physiological modelling of insulin and meal absorption significantly improved the performance
particularly in larger window size that enabled the model to better capture longer-term trends and temporal
dependencies.
1 INTRODUCTION
Type 1 Diabetes is a chronic metabolic condition
characterized by disrupted blood glucose homeostasis
due to the lack of endogenous insulin production
(Georga et al., 2012). As a result, subjects with type
1 diabetes must rely on exogenous insulin to
compensate for this deficiency (Oviedo et al., 2017).
To maintain blood glucose levels within the desired
range and to reduce the risk of complications such as
hypoglycaemia or hyperglycaemia, one must follow
a dietary guideline, administer insulin in right
amount, and proper exercise regimen (Zhu et al.,
2022). One promising technology for managing
blood glucose is continuous glucose monitoring
(CGM), which tracks glucose levels in real time and
is often integrated into automated insulin delivery
systems (Della Cioppa et al., 2023). These automated
systems, known as closed-loop control systems or
artificial pancreas, use predictive models to forecast
future blood glucose levels and adjust insulin delivery
accordingly.
Thus, accurate prediction of blood glucose levels
is vital for management of blood glucose. The use of
mathematical equations that represent the body’s
blood glucose dynamics to make accurate predictions
require detailed knowledge of individual physiology,
such as a person’s glucose absorption rates, insulin
sensitivity, and how their body handles glucose
during activities like eating or fasting (Oviedo et al.,
2017) (Woldaregay et al., 2019). They also depend on
a large number of pre-set parameters, requiring
extensive clinical and personal data about the
individual. On the other hand, data-driven models
focus more on self-monitored historical data such as
past glucose levels, insulin doses, and food intake and
not require as much detailed physiological
information (Woldaregay et al., 2019). While this
approach is easier to implement as it requires less
knowledge about the individual’s specific
physiology, it has limitations. These models are often
less consistent with actual physiological processes
and are of a black box nature. Also, many tend to lose
accuracy over longer timeframes (Ghimire et al.,
2024).
Previous study (Pawar et al., 2021) suggests that
combining simplified physics-based models with
neural network architecture can lead to improved
984
Ghimire, S., Celik, T., Gerdes, M. and Omlin, C. W.
Physiology-Guided Blood Glucose Predictive Model Using Minimal Blood Glucose Dynamics.
DOI: 10.5220/0013395200003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 984-990
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
predictive accuracy and reduced uncertainty. In line
with this principle, this paper investigates a hybrid
modelling paradigm. The hybrid model integrates the
subcutaneous insulin absorption kinetics and the meal
absorption dynamics within the predictive model for
blood glucose levels, as opposed to relying on
complex and data intensive full mathematical models.
It combines the strengths of physiological models
(which reflect real glucose dynamics) with machine
learning (ML) techniques (which can handle large
amounts of historical data). The primary aim is to
obtain more accurate blood glucose predictions that
remain consistent with physiological principles and
can be interpreted. Additionally, it intends to provide
a better understanding of its potential strength for
future expansion.
The remainder of this article is organized as
follows. The existing works are discussed in section
2. The description of the proposed method is
presented in section 3. Section 4 covers the data sets
used, data processing, and experiments setup,
including the evaluation metrics. In Section 5,
experimental results and analysis are provided.
Finally, the article is concluded in Section 6.
2 RELATED WORKS
Several studies (Bertachi et al., 2018; Contreras et al.,
2018; Contreras et al., 2017; Erdős et al., 2023; Georga
et al., 2012; Mougiakakou et al., 2006; Sun et al., 2020)
have developed models, integrating mathematical
models of blood glucose dynamics with data-driven
models. Study (Georga et al., 2012) enhanced
performance of the Support Vector Regression (SVR)
model by incorporating additional features such as the
rate of glucose appearance from meal intake (Ra),
plasma insulin levels (Ip), and cumulative glucose
appearance over a specific period (SRa). These
features, obtained from meal and insulin models of
blood glucose dynamics, serve as inputs, provide a
straightforward way to incorporate domain knowledge
into learning models. In a similar approach, study
(Mougiakakou et al., 2006) included plasma insulin
and glucose appearance rate from meals as input
features through insulin kinetics and meal models,
respectively. Additional features such as insulin on
board i.e., the remaining active insulin, carbohydrates
on board i.e., remaining carbs yet to be converted to
glucose, glucose appearance rate from carbs, rate of
glucose appearance in the blood from guts, and activity
on board were employed in studies (Bertachi et al.,
2018; Contreras et al., 2018; Contreras et al., 2017; Sun
et al., 2020) to refine ML models. Additionally, study
(Erdős et al., 2023) proposed a hybrid method that
sequentially combines predictions from both
mathematical and ML models, where the ML model
predicts residuals based on phenotypic features, which
are then subtracted from the predictions made by
mathematical model. The results demonstrated that the
personalized physiological models consistently
outperformed the data-driven and the hybrid model
approaches. Personalized physiological models may
inherently capture individualized critical processes and
features that data-driven models struggle to
individualize or interpret effectively. Although these
studies have shown improvements in performance
through the integration of various physiological
knowledge, they fail to analyse the potential of
integration across different contexts, a gap that is
addressed in this study.
3 PROPOSED METHOD
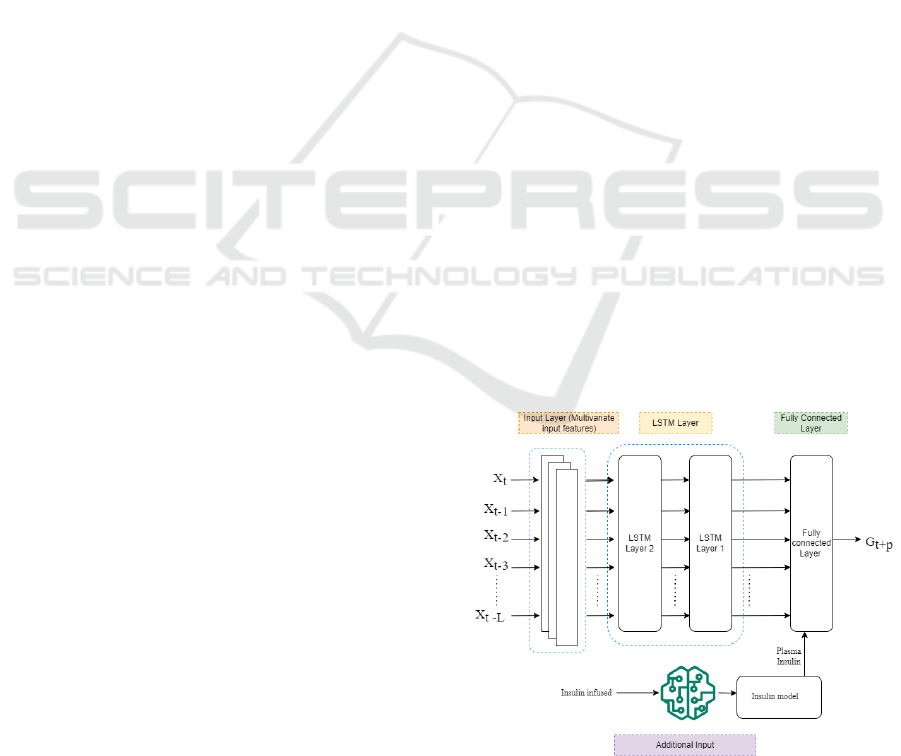
To explore the potential of the physiological inputs, a
straightforward model architecture was designed,
consisting of two Long short-term memory (LSTM)
layers with 64 units each, followed by a fully
connected layer with 64 neurons as the final layer, as
illustrated in Figure 1. LSTM networks are a
specialized type of recurrent neural networks (RNNs)
designed to effectively capture long-term temporal
dependencies in sequential data (Aliberti et al., 2019),
comprising memory cells, input, forget, and output
gates that dynamically manages information flow,
preserving the essential patterns and insights over
long sequences, enhancing their effectiveness for
time series prediction (Afsaneh et al., 2022).
Figure 1: System architecture of the proposed hybrid
model.
Physiology-Guided Blood Glucose Predictive Model Using Minimal Blood Glucose Dynamics
985
Different configurations of this model are used to
assess their potential performance. Predictive model
incorporating physiological inputs (insulin plasma (IP)
and carb on board (COB) including glucose (G)) is
labelled as PM, while the model without physiological
input is referred to as W-PM, that includes bolus (Bo),
basal (B), and carb (C) including G. The models are
further divided into input mode and output mode: in
input mode, the final layer receives no additional
inputs, while in output mode, an additional input
feature, IP, is included in the final layer.
When bolus insulin is administered, it doesn’t
instantly lower blood glucose; rather, it enters the
bloodstream, peaks after a certain time, and gradually
dissipates (Lehmann & Deutsch, 1992). Based on
similar physiology, the assumption is that if ML
model incorporates insulin dynamics that provides a
temporal representation of insulin’s effect, reflecting
how insulin concentration changes over time as is
seen in the body’s physiological process, it allows for
a more nuanced understanding of how blood glucose
will respond. For a simple bolus-only ML model that
considers only the dose and the timing of insulin
administration, however, is challenging to capture
how insulin gradually influences blood glucose.
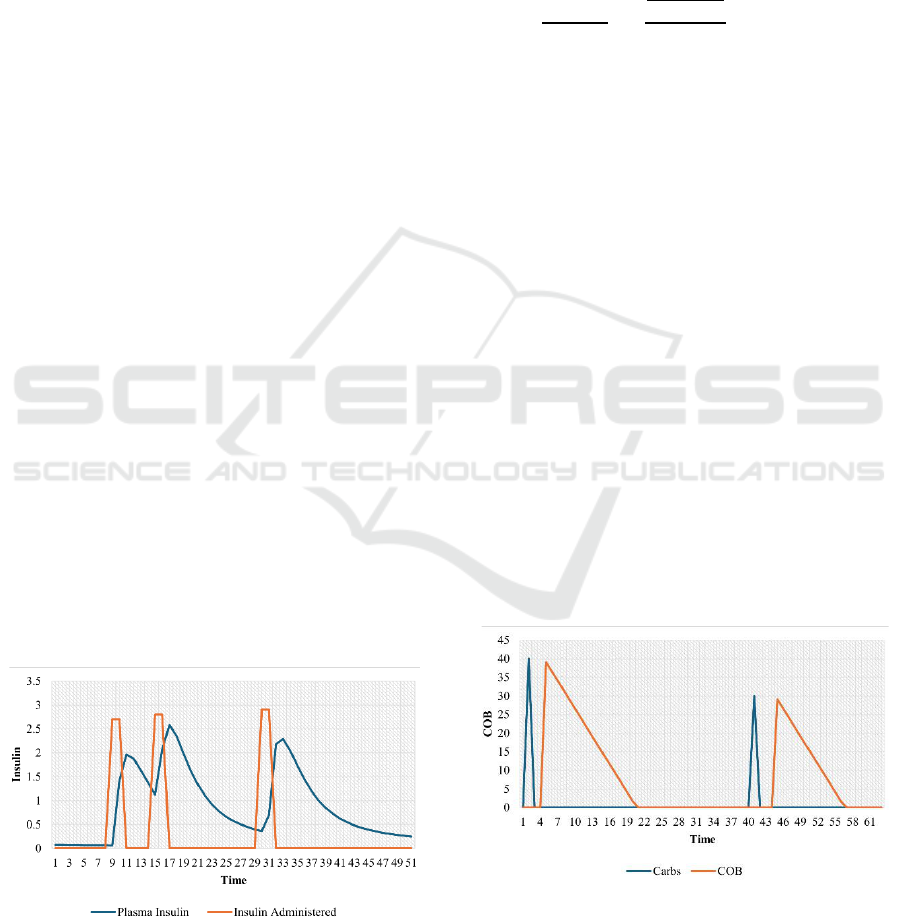
Figure 2 depicts the bolus administered dose (Marling
& Bunescu, 2020) and its timing curve along with the
insulin dynamics following the insulin dose, where
administered bolus is observed at a specific time step,
while the insulin dynamics profile exhibits a gradual
decay of insulin over time. It illustrates how insulin
levels rise and fall, giving a clear indication of how
glucose levels might influence. Additionally, giving a
second bolus dose before the previous dose has fully
dissipated results in cumulative insulin effects in the
bloodstream. By modelling plasma insulin, it is
possible to track these overlapping, enabling precise
predictions with increased insulin activity.
Figure 2: Insulin decay curve given by the physiological
insulin model.
Thus, based on this physiology, the proposed
model architecture incorporates the insulin dynamics
to provide a temporal representation of insulin’s
effect on how insulin concentration changes over
time, where we utilized the Lehmann model
(Lehmann & Deutsch, 1992) that aligns with the
physiological process of insulin absorption dynamics
as given in equation (1):
(1)
where, is plasma insulin concentration,
is the
first-order rate constant of insulin elimination, and
is the volume of insulin distribution, is the time
elapsed from the injection,
is the time at which
50% of the dose, D, has been absorbed and is a
preparation-specific parameter defining the insulin
absorption pattern of the different types of insulin.
The insulin dynamics that give plasma insulin is
based on rapid-acting insulin, which is known to have
an immediate effect on reducing blood glucose levels
with fast absorption and action. This makes it
particularly relevant for scenarios where quick
glucose adjustments are needed after a meal.
In addition to this, instead of using the discrete
carbohydrate data into the model, the carbohydrate on
board based on the meal intake is estimated to convert
it to a time action profile of absorption, similar to the
insulin dynamics as given by (2) (Dave et al., 2021).
It estimates the amount of carbs not yet appeared in
blood glucose. The curves in Figures 3 show how the
carbs decay over time.
(2)
Figure 3: The carbohydrate decay curve is given by the
physiological meal model.
HEALTHINF 2025 - 18th International Conference on Health Informatics
986
Here,
denote the time at which amount of
carbohydrates taken.
is the carbohydrate
absorption rate, which is 0.5g/min after initial delay
(
) of 15min and is the current time. These time
action profiles of insulin and carbs are fed into the PM
model as additional inputs along with glucose in
initial or final layer as shown in Figure 1. The plasma
insulin in the output mode is included in the final
layer of the model as shown in Figure 1, to act as a
corrective mechanism for the model to correct the
prediction at the last minute. This architecture implies
that the model first makes the prediction using input
features and then applies correction based on the
amount of insulin present or absorbed.
4 EXPERIMENTS
4.1 Datasets
The models were trained and tested using OhioT1DM
(Marling & Bunescu, 2020) dataset that encompasses
eight weeks of data from twelve patients with type-I
diabetes. It involves seven males and five females
within the age range of 20 to 80 years. The dataset
contains blood glucose measurements recorded every
five minutes, comprising total 166532 samples, using
Medtronic Enlite CGM sensors, along with bolus and
basal insulin administered via Medtronic 530G
insulin pumps at regular intervals. Also, the dataset is
supplemented by other daily events such as meal and
exercise info reported by the patients via a
smartphone app or fitness band. The proposed study
explores the data on blood glucose levels, insulin and
carbohydrate intake. The OhioT1DM dataset was
originally provided with separate training and testing
data for each of the 12 patients, which were directly
used for model training and evaluation. The training
data was further divided into an 80% training subset
and a 20% validation subset. The model, trained on
the training subset, was evaluated on the test set,
which consisted of new, unseen data.
4.2 Experimental Setup
To address missing data and standardize it,
resampling and normalization were applied. The
dataset was resampled to align all signals to a uniform
5-minute sampling grid. Data with consecutive gaps
exceeding 30 minutes were excluded to prevent
inaccuracies in prediction trajectories. Missing values
were imputed using linear interpolation; however, no
imputation was performed on the test set.
Standardized normalization was applied to scale the
data to a mean of zero and a standard deviation of one.
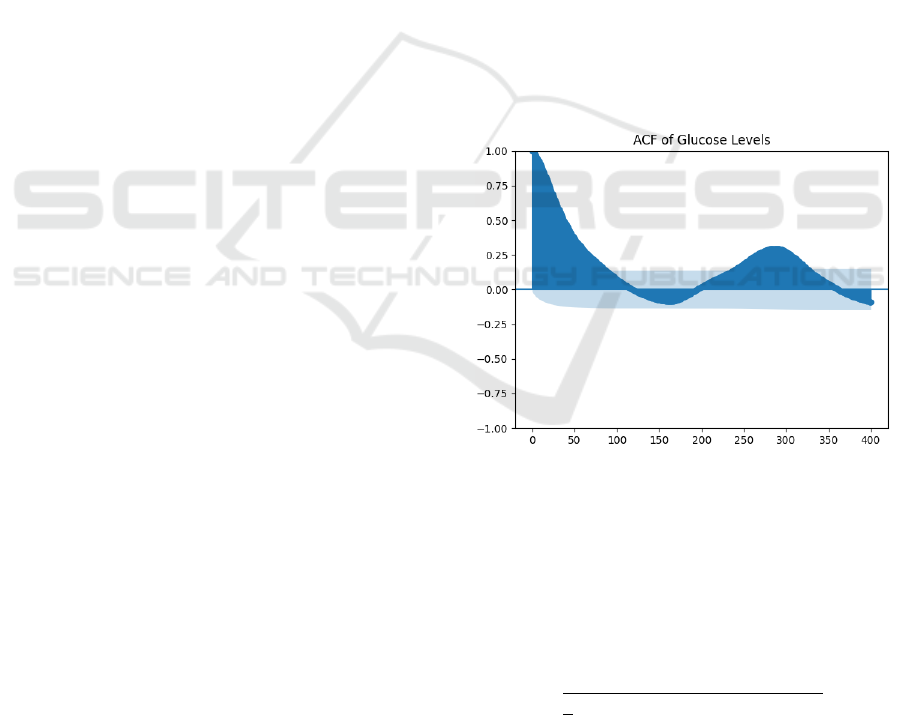
Additionally, to investigate the relationship
between blood glucose levels and their prior values
and to find the right window size, the autocorrelation
function (ACF) (Semmlow, 2012) was utilized. As
ACF describes how well a signal correlates with
adjacent part of itself, the plot of the ACF on blood
glucose levels revealed a strong correlation between
the current glucose levels and glucose levels from the
previous day as shown in Figure 4. Based on this
analysis, a sliding window of 24 hours was used
instead of a 2-hour historical data window to predict
blood glucose levels 30 minutes in advance, as
opposed to the literatures (Cui et al., 2021; Ghimire
et al., 2024; Oviedo et al., 2017). The model was
trained for 100 epochs with 10 repetitive runs using
Huber loss (Tong, 2023) as the cost function. The
adaptive moment estimation (Adam) optimizer was
employed to minimize the loss function.
Optimization parameters included a learning rate
decay of 0.1, a decay patience of 10, and early
stopping patience of 30.
Figure 4: Auto-correlation function of blood glucose with
its previous values of 24hrs.
4.3 Evaluation Metrics
Model performance was assessed using the root mean
square error (RMSE), a standard metric that
quantifies the similarity between predicted and actual
blood glucose levels, defined as follows:
(3)
Here, represents the number of test samples,
denotes the actual blood glucose levels,
is the
predicted glucose level, and PH stands for the
Physiology-Guided Blood Glucose Predictive Model Using Minimal Blood Glucose Dynamics
987
prediction horizon—the time frame for which blood
glucose levels are forecasted. A lower RMSE value
indicates better model performance in glucose
prediction. For validation, we employed a 30-minute
prediction horizon (PH), implies that the model
predicts blood glucose levels 30 minutes ahead.
5 EXPERIMENTAL RESULTS
AND DISCUSSION
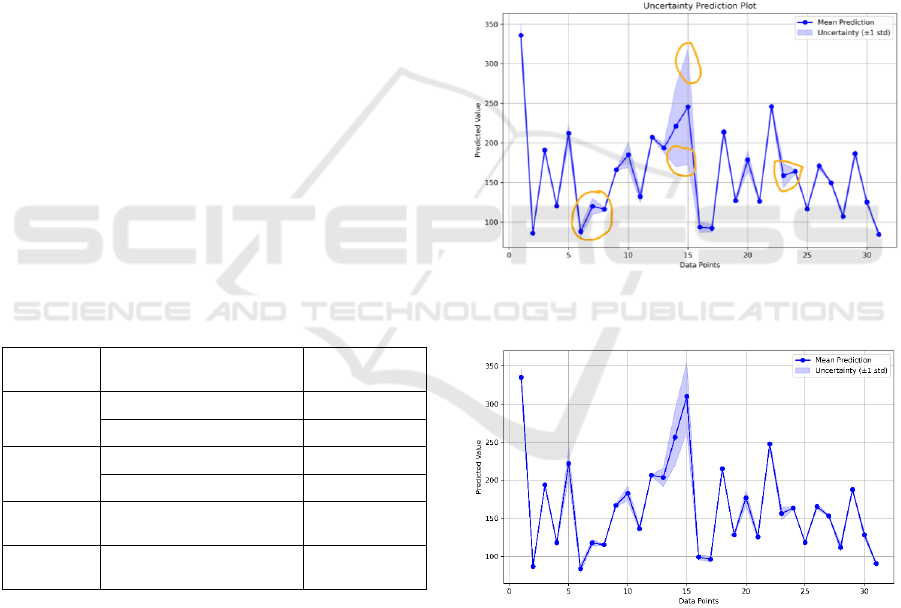
Table 1 presents the results for the proposed model
with a 30-minute PH across various input feature
combinations, both with and without a physiological
model. The results indicate that the PM model
attained a lower RMSE than the W-PM model,
demonstrating the significance of the use of
physiological inputs. While adding insulin plasma in
the final layer did not reduce RMSE in either
approach, it enhanced the prediction certainty
compared to models without plasma insulin, as is also
illustrated in Figures 5 and 6 with highlights. No
significant difference in RMSE was observed
between input and output modes within the PM
model. Overall, incorporating physiological
dynamics led to a performance improvement of over
10%, and adding physiological dynamics in the final
layer contributed further to prediction certainty.
Table 1: Summary of the RMSE(Std) result for different
combinations of input features.
Predictive
Models
Input Mode (in features)
RMSE(Std)
W-PM
(24hrs)
Output (G, BO, C, B) (IP)
20.154 (1.32)
Input (G, BO, C, B)
20.142 (2.08)
PM (24hrs)
Output (G, CO) (IP)
17.606 (0.13)
Input (G, CO, IP)
17.526 (0.159)
W-PM (2hrs)
Input (G, BO, C, B)
19.235 (0.135)
PM (2hrs)
Input (G, CO, IP)
19.437 (0.233)
PM: Physiological Model, W-PM: Without Physiological
Model, G: Glucose level, BO: Bolus insulin, C:
Carbohydrate, B: Basal insulin, CO: Carbohydrate on
Board and IP: Plasma Insulin
From the results, it is evident that using
physiological models offers no significant
improvement when a shorter 2-hour window is
employed. The potential reason for this is the
continuous data sampled at 5-minute intervals using
physiological modelling for insulin and meal,
successfully created smoother transitions between
data points, allowing the models to see gradual
changes rather than abrupt shifts. With a 24-hour
window, this continuous representation enabled the
model to better capture longer-term trends and
temporal dependencies. In contrast, for the discrete
insulin and carbs data, model struggled to identify
patterns over longer windows due to the limited
number of data points within each window, which
constrained the model’s ability to learn these trends
effectively. However, with a 2-hour window, the
difference between the continuous and discrete is
minimal and there is enough overlap between the
neighbouring data points within the window, making
the abrupt changes in the discrete insulin and carbs
data less impactful on model performance.
Figure 5: Uncertainty prediction plot of input mode of W-
PM model.
Figure 6: Uncertainty prediction plot of output mode of PM
model.
To further explain the output of the predictive
model, Shapley additive explanation (SHAP), a
model agnostic explainability approach is utilized.
SHAP plots as shown in Figures 7 and 8 for both
models highlight that blood glucose is the most
influential feature in driving the model's decisions,
with carbs (or COB in the case of the PM model)
HEALTHINF 2025 - 18th International Conference on Health Informatics
988
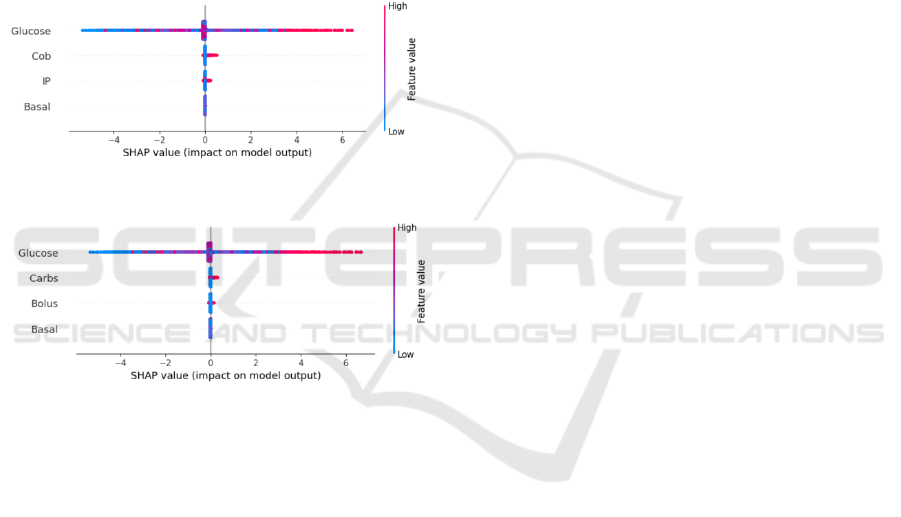
being the second most important. Bolus and basal
have a minimal to no impact on the output
predictions, though bolus shows a slightly higher
influence. A higher SHAP value for COB compared
to carbs indicates that the PM model assigns
significant importance to COB in predicting future
blood glucose levels. This also suggests that COB has
a notable positive effect on predictions, aligning with
the physiological process where an increase in COB
leads to an increase in future blood glucose levels,
demonstrating the prediction to be consistent with the
physiology. This observation supports the superior
performance of the PM model, which benefits from
the inclusion of physiological inputs compared to the
W-PM model.
Figure 7: Summary plot of SHAP analysis of PM model.
Figure 8: Summary plot of SHAP analysis of W-PM model.
6 LIMITATIONS
Despite potential results, this study has some
limitations. The study modelled the physiological
process using only bolus insulin, excluding basal
insulin, which plays a crucial role in blood glucose
regulation. Also, as the study focused on a simplified
physiological model, incorporating more complex
physiological processes could provide deeper insights
and improve prediction accuracy. In addition, the
SHAP analysis highlighted which input features
contribute most to the model’s predictions, revealing
associations identified by the model, but cannot
provide the underlying causal mechanisms.
Therefore, for drawing causal conclusions additional
analysis such as counterfactual analysis can
potentially be beneficial.
7 CONCLUSIONS
This study introduced a straightforward integration of
a physiological model with a data-driven approach for
predicting blood glucose levels. The findings
demonstrated that this hybrid model, incorporating a
simple physiological model, has the potential to
enhance predictive performance, also increase
certainty when integrated into the final layer.
Moreover, the analysis highlights that incorporating
the physiological process of gradual change
demonstrates its significance with larger window
sizes. This inclusion notably improved model
performance by preserving temporal dynamics that
would otherwise be lost in discrete data lacking a
physiological basis. Additionally, blood glucose is
identified as the most influential contributor for
prediction output, followed by COB, demonstrating
the importance of physiological dynamics.
Given the study's demonstrated potential for
physiological integration, future work could focus on
incorporating complex physiological modelling into
the model to enhance its representation of
physiological processes, predictive accuracy and
explainability.
REFERENCES
Afsaneh, E., Sharifdini, A., Ghazzaghi, H., & Ghobadi, M.
Z. (2022). Recent applications of machine learning and
deep learning models in the prediction, diagnosis, and
management of diabetes: a comprehensive review.
Diabetology & Metabolic Syndrome, 14(1), 1-39.
Aliberti, A., Pupillo, I., Terna, S., Macii, E., Di Cataldo, S.,
Patti, E., & Acquaviva, A. (2019). A multi-patient data-
driven approach to blood glucose prediction. IEEE
ACCESS, 7, 69311-69325.
Bertachi, A., Biagi, L., Contreras, I., Luo, N., & Vehí, J.
(2018). Prediction of Blood Glucose Levels And
Nocturnal Hypoglycemia Using Physiological Models
and Artificial Neural Networks. KDH@ IJCAI,
Contreras, I., Bertachi, A., Biagi, L., Vehí, J., & Oviedo, S.
(2018). Using Grammatical Evolution to Generate
Short-term Blood Glucose Prediction Models. KDH@
IJCAI,
Contreras, I., Oviedo, S., Vettoretti, M., Visentin, R., &
Vehí, J. (2017). Personalized blood glucose prediction:
A hybrid approach using grammatical evolution and
physiological models. PLoS ONE, 12(11), e0187754.
Cui, R., Hettiarachchi, C., Nolan, C. J., Daskalaki, E., &
Suominen, H. (2021). Personalised short-term glucose
prediction via recurrent self-attention network. 2021
IEEE 34th International Symposium on Computer-
Based Medical Systems (CBMS),
Physiology-Guided Blood Glucose Predictive Model Using Minimal Blood Glucose Dynamics
989

Dave, D., DeSalvo, D. J., Haridas, B., McKay, S., Shenoy,
A., Koh, C. J., Lawley, M., & Erraguntla, M. (2021).
Feature-based machine learning model for real-time
hypoglycemia prediction. Journal of Diabetes Science
and Technology, 15(4), 842-855.
Della Cioppa, A., De Falco, I., Koutny, T., Scafuri, U., Ubl,
M., & Tarantino, E. (2023). Reducing high-risk glucose
forecasting errors by evolving interpretable models for
type 1 diabetes. Applied soft computing, 134, 110012.
Erdős, B., van Sloun, B., Goossens, G. H., O’Donovan, S.
D., de Galan, B. E., van Greevenbroek, M. M.,
Stehouwer, C. D., Schram, M. T., Blaak, E. E., &
Adriaens, M. E. (2023). Quantifying postprandial
glucose responses using a hybrid modeling approach:
Combining mechanistic and data-driven models in The
Maastricht Study. PLoS ONE, 18(7), e0285820.
Georga, E. I., Protopappas, V. C., Ardigo, D., Marina, M.,
Zavaroni, I., Polyzos, D., & Fotiadis, D. I. (2012).
Multivariate prediction of subcutaneous glucose
concentration in type 1 diabetes patients based on
support vector regression. IEEE Journal of Biomedical
and Health Informatics, 17(1), 71-81.
Ghimire, S., Celik, T., Gerdes, M., & Omlin, C. W. (2024).
Deep learning for blood glucose level prediction: How
well do models generalize across different data sets?
PLoS ONE, 19(9), e0310801.
Lehmann, E., & Deutsch, T. (1992). A physiological model
of glucose-insulin interaction in type 1 diabetes
mellitus. Journal of biomedical engineering, 14(3),
235-242.
Marling, C., & Bunescu, R. (2020). The OhioT1DM dataset
for blood glucose level prediction: Update 2020. CEUR
workshop proceedings,
Mougiakakou, S. G., Prountzou, A., Iliopoulou, D., Nikita,
K. S., Vazeou, A., & Bartsocas, C. S. (2006). Neural
network based glucose-insulin metabolism models for
children with type 1 diabetes. 2006 International
Conference of the IEEE Engineering in Medicine and
Biology Society,
Oviedo, S., Vehí, J., Calm, R., & Armengol, J. (2017). A
review of personalized blood glucose prediction
strategies for T1DM patients. International journal for
numerical methods in biomedical engineering, 33(6),
e2833.
Pawar, S., San, O., Aksoylu, B., Rasheed, A., & Kvamsdal,
T. (2021). Physics guided machine learning using
simplified theories. Physics of Fluids, 33(1).
Semmlow, J. (2012). Autocorrelation Function. Retrieved
October from
https://www.sciencedirect.com/topics/engineering/auto
correlation-function
Sun, X., Rashid, M. M., Sevil, M., Hobbs, N., Brandt, R.,
Askari, M.-R., Shahidehpour, A., & Cinar, A. (2020).
Prediction of Blood Glucose Levels for People with
Type 1 Diabetes using Latent-Variable-based Model.
KDH@ ECAI, 20, 115-119.
Tong, H. (2023). Functional linear regression with Huber
loss. Journal of Complexity, 74, 101696.
Woldaregay, A. Z., Årsand, E., Walderhaug, S., Albers, D.,
Mamykina, L., Botsis, T., & Hartvigsen, G. (2019).
Data-driven modeling and prediction of blood glucose
dynamics: Machine learning applications in type 1
diabetes. Artificial intelligence in medicine, 98, 109-
134.
Zhu, T., Li, K., Herrero, P., & Georgiou, P. (2022).
Personalized blood glucose prediction for type 1
diabetes using evidential deep learning and meta-
learning. IEEE Transactions on Biomedical
Engineering, 70(1), 193-204.
HEALTHINF 2025 - 18th International Conference on Health Informatics
990
